|
|
| Chinese Scientists Reveal Replication Mechanism of MPXV DNA Polymerase holoenzyme |
Monkeypox virus (MPXV) belongs to the Orthopoxvirus genus of the Poxviridae family, which also includes the variola virus that causes smallpox and has killed millions of humans in recorded history. The World Health Organization announced the monkeypox as a public health emergency of international concern in July 2022, and prophylactic and therapeutic measures are in urgent need. The MPXV is a large double-stranded DNA virus that replicates exclusively in the cytoplasm of the infected cells. After entry, the virus initiates early gene transcription events, and viral DNA synthesis occurs at perinuclear sites called viral factories. The MPXV replicative holoenzyme contains catalytic polymerase F8, a heterodimeric processivity factor consisting of A22 and uracil-DNA glycosylase E4, which is an ideal target for drug development. Although previous genetic, biochemical, and structural studies on the Vaccina virus (VACV) core replication machinery have advanced our understanding of poxvirus DNA replication, we are still awaiting a reliable high-resolution structure of the replicating state of the Orthopoxvirus polymerase holoenzyme, and the mode of operation of the processivity factor needs to be elucidated. Recently, a joint team led by Dr. George Fu Gao and Dr. Yi Shi from Institute of Microbiology, Chinese Academy of Sciences (CAS), have recently investigated the working mechanism of MPXV polymerase. In this study, they determined the structure of MPXV DNA polymerase holoenzyme in replicating state. The structure of the holoenzyme/DNA complex contains one F8, one A22, one E4, and the primer/template DNA, as well as an incoming dTTP substrate in a closed conformation. The F8 and E4 resemble the counterparts of VACV. A22 can be divided into three domains: the A22 NTD, middle domain (Mid), and C-terminal domain (CTD), in which the NTD and CTD bind with E4 and F8, respectively. The Mid displays high structural similarities with the DNA ligases, but does not possess ordinary ligase activity like T4 ligase (Fig. 1). Compared with the structures of adenylation domains from the T4 and ASFV ligases, the putative ligase active site of the A22 Mid is replaced by hydrophobic and negatively charged residues, which may prevent the binding of ATP substrate. Therefore, A22 may comprise a degenerative ligase domain acting simply as a flexible linker. 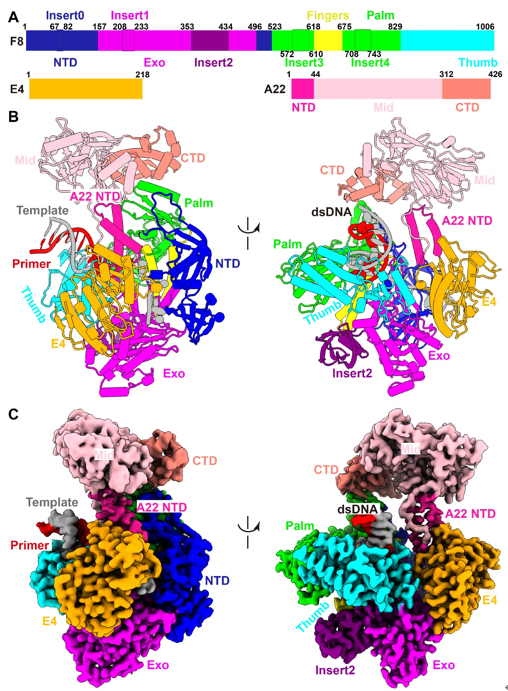 Fig. 1. Overall structure of the replicating MPXV DNA polymerase holoenzyme. 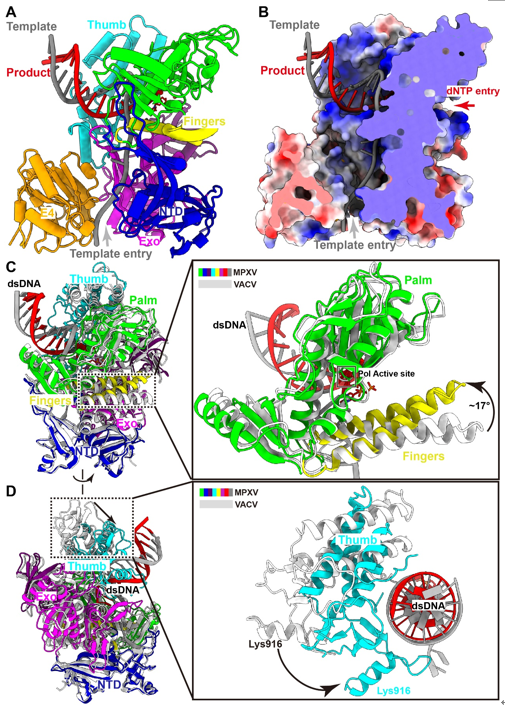 Fig. 2. Conformational changes from apo to replicating states.
MPXV polymerase has a similar DNA binding mode to other B-family DNA polymerases from different species. Compared with the structure of VACV polymerase in apo state, the finger domain rotates toward the palm domain by approximately 17° in the replicating state. This rotation drags the positively charged Arg634 and Lys661 of the fingers domain closer to the active site, where they can interact with the triphosphate group of incoming dNTP. The thumb domain also makes a significant rotation to wrap around the primer-template DNA duplex on its minor groove side. The DNA duplex is accommodated in a positively charged groove of the thumb domain, as observed in other B-family DNA polymerases (Fig. 2). For B-family DNA polymerases, proliferating cell nuclear antigen (PCNA) or PCNA-like proteins are required for high processivity. However, for poxviruses, including MPXV and VACV, no homologous PCNA-like proteins have been identified in the viral genome. Instead, the poxvirus-specific A22-E4 heterodimer is responsible for the high processivity of DNA replication. In this study, they also demonstrated that F8 polymerase alone dissociated from the primer-template DNA after incorporating less than 14 nt, while the addition of the A22-D4 heterodimer conferred processivity to F8 in a concentration-dependent manner. The E4 cofactor interacts with the Exo of F8 polymerase, and together with the NTD of F8, they form a closed-ring channel to encircle the single-stranded template DNA and prevent the dissociating from holoenzyme complex, proposed as “forward sliding clamp”. In contrast, in the yeast DNA polymerase complex, the Exo and NTD form an open semicircular channel to accommodate the single-stranded template DNA, and the trimeric PCNA ring encircles the template-product DNA duplex, recognized as “backward sliding clamp” (Fig. 3). This architectural difference between MPXV and yeast polymerase complexes is responsible for the different processivity mechanisms during DNA replication events. In conclusion, these findings reveal the mechanism of the MPXV genome replication and may guide the development of anti-poxvirus drugs. 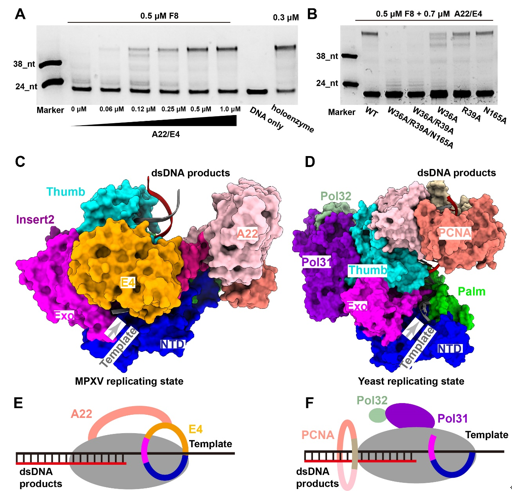 Fig. 3. The unique mechanism of A22-E4 heterodimer promoting processivity of MPXV DNA polymerase. Full text links: https://www.science.org/doi/10.1126/science.ade6360
|
|
|
|
|
|
