The outbreak of COVID-19 has developed into a global pandemic affecting almost all countries in the world. As of 1st June 2020, more than 6,300,000 human infections have been reported worldwide, including over 370,000 deaths, according to the statistics by Johns Hopkins University. Currently, there are no specific drugs or vaccines available for this newly emerging virus.
COVID-19 is caused by a novel coronavirus named Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), which belongs to the family of Coronaviridae, a group of positive-sense RNA virus with a broad host-spectrum. So far, a total of seven human-infecting coronaviruses have been identified.
Among them, SARS-CoV-2 displays the highest similarity in genome sequence to SARS-CoV which emerged in 2002-2003. Both viruses utilize the same host receptor angiotensin-converting enzyme 2 (ACE2) for cell entry and cause respiratory symptoms that may progress to severe pneumonia and lead to death.
However, as compared to SARS-CoV, SARS-CoV-2 shows a much higher transmission efficiency and a lower mortality rate. Most of the infections result in mild symptoms and a substantial number of asymptomatic infection cases have also been reported.
The replication of coronavirus is operated by a set of non-structural proteins (nsps) encoded by its genome, which are initially translated as polyproteins followed by proteolysis cleavage to generate mature viral proteins. These proteins assemble into a multi-subunit polymerase complex to mediate the transcription and replication of viral genome.
Among them, nsp12 is the catalytic subunit with RNA-dependent RNA polymerase (RdRp) activity. The nsp12 itself is capable of conducting polymerase reaction with extremely low efficiency, whereas the presence of nsp7 and nsp8 cofactors remarkably stimulates its polymerase activity. The nsp12-nsp7-nsp8 subcomplex is thus defined as the minimal core components for mediating coronavirus RNA synthesis.
To achieve the complete transcription and replication of viral genome, several other nsp subunits are required to assemble into a holoenzyme complex, including the nsp10, nsp13, nsp14 and nsp16, for which the precise functions for RNA synthesis have not been well understood.
On 30th May 2020, SHI Yi’s Group at Institute of microbiology, Chinese Academy of Science reported the 3D structure and enzymatic properties of the core polymerase complex from COVID-19 virus in Cell Reports.
This complex consists of the nsp12 catalytic subunit bound with a copy of nsp7 and two asymmetric nsp8 cofactors. The structure highly resembles the counterpart of SARS-CoV with conserved motifs for all viral RNA-dependent RNA polymerase, and suggests the mechanism for catalysis activation by cofactors. These findings provide important basis for developing antiviral drugs that inhibit RNA synthesis by targeting the viral polymerase complex.
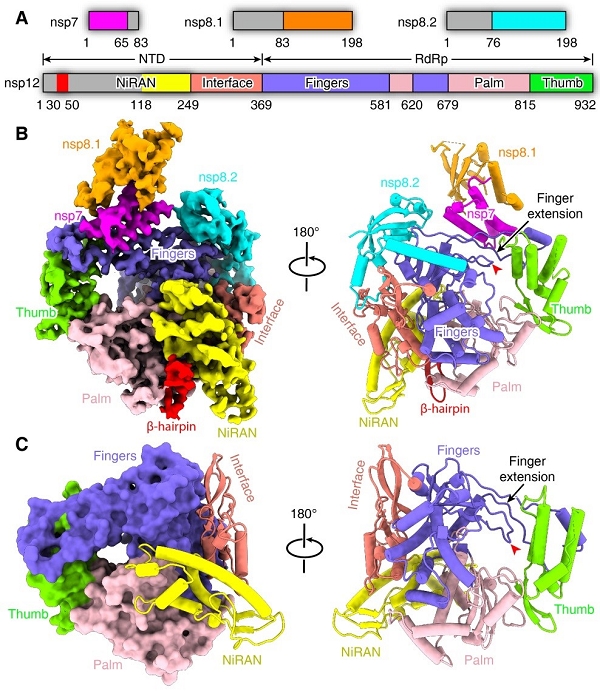
Figure 1. Overall structure of SARS-CoV-2 core polymerase complex (Image by SHI Yi’s group)
Interestingly, in vitro enzymatic activity assays reveals that the core polymerase complex of SARS-CoV-2 displays a significantly reduced efficiency for RNA synthesis as compared to that of SARS-CoV.
Moreover, the thermostability of individual subunits of SARS-CoV-2 core polymerase complex is also obviously lower than the counterparts of SARS-CoV. These observations may provide clues for the adaptive evolution of SARS-CoV-2 to enable efficient transmission among human populations.
Previous bioinformatic studies suggested that both SARS-CoV and SARS-CoV-2 may have originated from the same natural host-bats. SARS-CoV has not fully adapted to humans and ferret, but SARS-CoV-2 displays efficient human-to-human transmission capabilities, suggesting the better adaptation of SARS-CoV-2 for human hosts.
Moreover, the body temperature of humans is significantly lower than that of bats which could even rise above 40 ℃ during flight. The lower thermostability of SARS-CoV-2 polymerase subunits compared to SARS-CoV thus constitutes important evidence for the well adaptation of SARS-CoV-2 towards humans, and may further support the bat-origin of related coronaviruses.
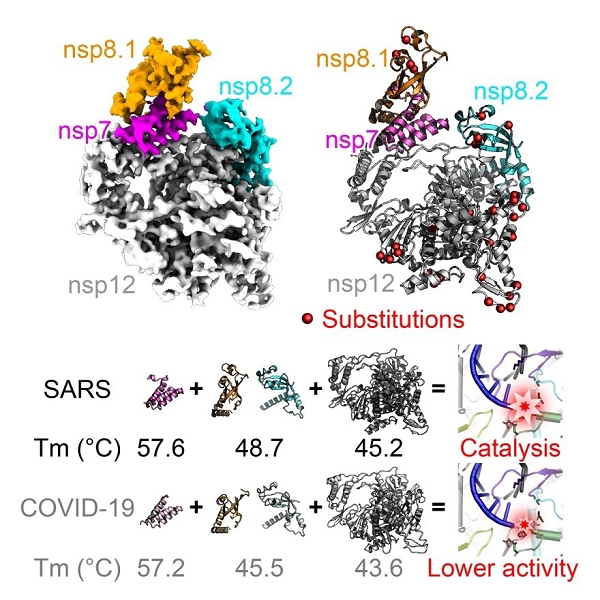
Figure 2. Compare of structural and biochemical properties of core polymerase from SARS-CoV-2 and SARS-Cov. (Image by SHI Yi’s group)
This work improves our understanding on the mechanism of RNA synthesis by SARS-CoV-2 and suggests new clues for the adaptive evolution of coronaviruses in favor of human hosts.
Paper link: https://www.cell.com/cell-reports/fulltext/S2211-1247(20)30754-3
