Streptococcus pneumoniae is the most common human respiratory pathogen, and β-lactam antibiotics have been employed to treat infections caused by S. pneumoniae for decades.
β-lactam resistance is steadily increasing in pneumococci and is mainly associated with the alteration in penicillin-binding proteins (PBPs) that reduce binding af?nity of antibiotics to PBPs.
However, the high variability of PBPs in clinical isolates and their mosaic gene structure hamper the predication of resistance level according to the PBP gene sequences.
A research group led by Prof. FENG Jie at Institute of Microbiology, Chinese Academy of Sciences developed a systematic strategy for applying supervised machine learning (SL) to predict AST of β-lactam antibiotic resistance. The published PBP sequences with minimum inhibitory concentration (MIC) values and the sequences from NCBI database without MIC values were served as labelled data and unlabeled data, respectively. The performances of SL models were evaluated by cross-validation: the labelled data set was randomly split into 80% training set and 20% test set 100 times.
This work demonstrated the highly variant amino acid loci (HVLs) are associated with antibiotic resistance using the unlabeled data in public database in NCBI (Figure).
Association between amino acid loci was demonstrated as an approach for feature reduction using only HVLs or sequence fragments during building machine learning models. The cefuroxime and amoxicillin resistance can be predicted well only using fragment from pbp2x (750 bp) and a fragment from pbp2b (750 bp), which allows one Sanger sequencing reaction to predicate the resistance phenotype.
Furthermore, the precision of predication model was evaluated by constructing the mutants containing the pbps from S. pneumoniae strains, of which genomes are available in NCBI database, and their phenotypes were predicated according to the model.
The model was tested by predicting resistance phenotypes of a local clinical strain collection. Both these approaches validated that the SL model could predicate the phenotype accurately.
Besides, a correlation between resistant phenotype and serotype and phylogenesis was revealed in more than 8000 S. pneumoniae strains available from NCBI database by applying the approach, which facilitates the understanding of the worldwide epidemiology of S. pneumonia.
Overall, this study established an effective genotypic AST approach for detecting β-lactam resistance levels in S. pneumoniae.
This study supported by the National Natural Science Foundation of China and the Beijing Municipal Science & Technology Commission was published in Briefings in Bioinformatics.
https://academic.oup.com/bib/advance-article/doi/10.1093/bib/bbz056/5512424
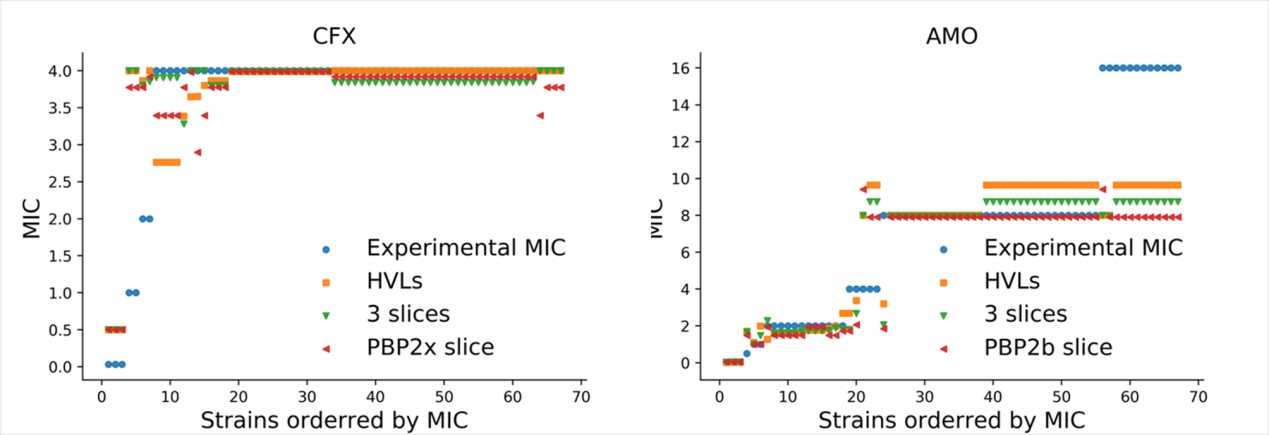
Figure Random forest model prediction on 62 strains. Experimental MIC: the MIC of 62 strains detected. HVLs: Highly Variant Locis were used to predict as input feature. 3 slices: 3 fragments captured from 3 PBP ends in series were used to predict as input feature. PBP2b slice: Fragment from PBP2b was used to predict as input feature. PBP2x slice: fragment from PBP2x was used to predict as input feature. CFX, cefuroxime; AMO, amoxicillin (Image by Prof. FENG’s group).
Contact:
Prof. FENG Jie
State Key Laboratory of Microbial Resources,
Institute of Microbiology, Chinese Academy of Sciences,
Beijing 100101
Email: fengj@im.ac.cn
