Chinese scientists discovered that trans-kingdom small RNAs (sRNAs) can be used to protect crops from infection of fungal pathogens. This RNAi-based mechanism has been successfully manipulated to protect cotton from infection of a soil-borne fungal pathogen Verticillium dahlia (V. dahliae), the causal agent of vascular wilt disease so-called “Cotton Cancer” in China.
As the world’s largest cotton producer, China yields 6-8 million tons cotton (30% of total world production) every year. However, high quality cotton cultivars are vulnerable to Verticillium wilt disease. Due to the long-term survival and vascular colonization of pathogen, the disease cannot be controlled by fungicides. How V. dahliae infects cotton was largely unknown and very limited resistance genes can be used for cotton breeding. Thus, no efficient methods were used to protect this economically important crop. In 2015, over 50% cotton production bases in Xinjiang Province (the largest cotton growing area in China) were under pressure of Verticillium wilt disease. Farmers had to accept severe yield loss after giving the whole year’s effort.
In 2008, when Prof. GUO Hui-Shan, Principal Investigator of Institute of Microbiology, Chinese Academy of Sciences visited Xinjiang Province for the first time she was astonished by the huge area of infected cottons and desperate farmers. After the trip in Xinjiang she became interested in the process of cotton-V. dahliae infection, and led her research group searching for a novel strategy to target V. dahliae. Eight years of trials in the lab as well as in the field have demonstrated three major achievements in understanding Verticillium wilt disease.
Firstly, trans-kingdom sRNAs exist during cotton-V. dahliae interactions, and some target virulence genes of pathogens (http://www.nature.com/articles/nplants2016153). Secondly, a novel penetration structure “hyphopodium” helps V. dahliae to “break” plant cell, which determines successful fungal infection (http://dx.doi.org/10.1371/journal.ppat.1005793). Thirdly, and most importantly, cotton cultivars expressing specific sRNAs are resistant to Verticillium wilt disease (http://dx.doi.org/10.1016/j.molp.2016.02.008). These results have been published in the journals such as Nature Plants, PLoS Pathogens and Molecular Plant, respectively, contributing not only to scientific community but also to crop disease management.
The route to the achievement was obviously not easy. Before the project started, Prof. GUO’s group already had strong background on RNAi mechanism in model plants but lack experience on crops especially cotton. Since host-induced gene silencing (HIGS) had been reported in plant-insect interaction, it raised an idea that HIGS might be an efficient strategy to inhibit hyphal growing in cotton.
Taking advantage of RNA & genome sequencing, Prof. GUO and her colleagues found that cotton sRNAs exist in V. dahliae hyphae retrieved from infected cotton, which means sRNAs naturally transmit from host to pathogens during cotton-V. dahliae interaction. Surprisingly, some transmitted sRNAs target fungal genes involved in pathogenicity and efficiently degrade gene transcripts. This phenomenon gives confidence to Prof. GUO. She believes that cotton would gain disease resistance by stably expressing sRNAs that target V. dahliae virulence genes.
“It was really time-consuming to generate the cottons stably expressing specific sRNAs, but we obtained them finally”, Prof. GUO said, “and luckily these cottons showed resistance to the wilt disease!” The new generated cottons show disease resistance not only in the lab but also in the field, in which the fungal populations are very diverse.
After six generations growing in Xinjiang Province, sRNA expressing cottons showed 22.25% more disease resistance in compared to the other cotton cultivars in the same cotton production base. It is quite enjoyable for Prof. GUO and colleagues to see the resistant cotton growing in the field, although it seems to be just the first step of disease management. More efficient fungal targets need to be found out based on the established systems, which can be also guidelines to other un-controlled crop diseases in the future.
The paper entitled “Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen” has been published online in Nature Plants (Zhang et al., 2016) with the Ph.D. student ZHANG Tao, ZHAO Yun-long and research assistant Dr. ZHAO Jian-Hua as co-first authors and Prof. GUO as corresponding author.
Their work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11040500) and the China Transgenic Research and Commercialization Key Special Project (2014ZX00800908B).
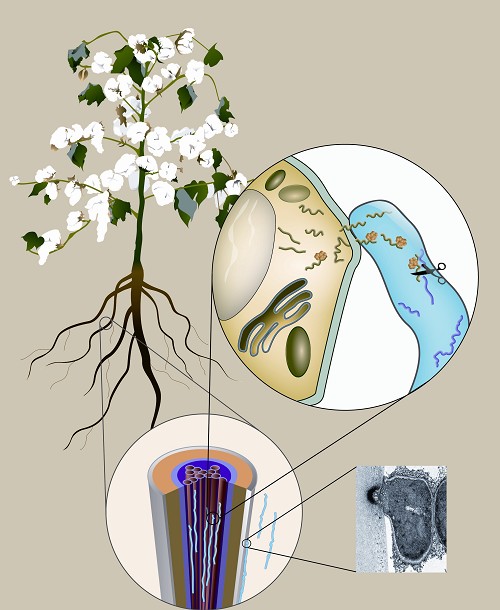
Figure legend: Trans-kingdom small RNAs target Verticillium dahliae genes during plant-microbe interaction. The soil-borne fungal pathogen V. dahliae (blue in the lower figure) develops infectious structure “hyphopodium” (Transmission electron microscopy image in the lower figure on the right) to penetrate and infects cotton roots and colonizes plant vascular tissues (red tubes in the lower figure). Small RNAs (green lines in the figure on the right) transmit from the plant cell to the colonized hyphae (blue in the figure on the right), targeting and “cutting”fungal gene transcripts (purple lines in the figure on the right). (Image by Prof. GUO’s group)
Contact
Prof. GUO Huishan
Institute of Microbiology, Chinese Academy of Sciences
E-mail: guohs@im.ac.cn
