The 2014 Ebola outbreak in West Africa has shocked the entire world, causing over 25,000 human infections with over 10,000 died. In September 2014, the Chinese government dispatched the China Mobile Laboratory Testing Team upon the request of the Sierra Leone government. From September to November, the Chinese teams detected over 3,000 samples with over 800 were tested Ebola virus-positive. On May 13th, 2015, Nature published online the latest Ebola research “Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone”, mainly accomplished by the Chinese scientists, with the aid of the Sierra Leonean Ministry of Health. The Member of the Chinese Academy of Science, professor George F. Gao of the Institute of Microbiology, Chinese Academy of Sciences acts as the co-leader of this project, designed and supervised the processes. He is also the co-corresponding author of this publication. Professor Di Liu of the Institute of Microbiology, Chinese Academy of Science, who is the co-first author of the paper, involved in analyzing the data, interpreting the results, and writing the paper.
In order to depict the virus transmission linkage/network in West Sierra Leone and to discern the phylodynamics of this novel EBOV, Chinese scientists have sequenced 175 full virus genomes. Through the comparison of the earlier published virus genomes, genetic diversity and evolutionary dynamics of the 2014 Ebola virus were concluded.
One of the marked findings is that the Ebola virus evolves at a steady evolutionary rate as it was. From an earlier dataset, Gire et al. estimated a faster evolutionary rate of the virus and posed the concerns that virus would become more harmful. This study with a combined dataset claims that virus did not evolve in a faster speed.
Another important finding is that the virus keeps becoming divergent as expect, and a total of 440 single nucleotide substitutions were discovered. However, there is no evidence that Ebola accumulates harmful mutations as it spread through Sierra Leone.
Moreover, scientists noticed some patients existed serial nucleotide substitutions, which was possibly occurred via same evolutionary events. The mechanisms underlying these observations need further investigations.
In general, this study provided new insights into the Ebola virus evolution in West Africa, and knowing the genetic diversity and evolutionary dynamics would greatly aid the public to fight against Ebola.
The study was accomplished by scientists from top Chinese institutes and with the aid of the Sierra Leonean Ministry of Health. Prof. George Fu Gao and Dr. LIU Di from the Institute of Microbiology, Chinese Academy of Sciences are co-corresponding author and co-first author respectively.
Nature published online this latest Ebola research (http://dx.doi.org/10.1038/nature14490), with a comment “Latest Ebola data rule out rapid mutation”(http://www.nature.com/news/latest-ebola-data-rule-out-rapid-mutation-1.17554). .
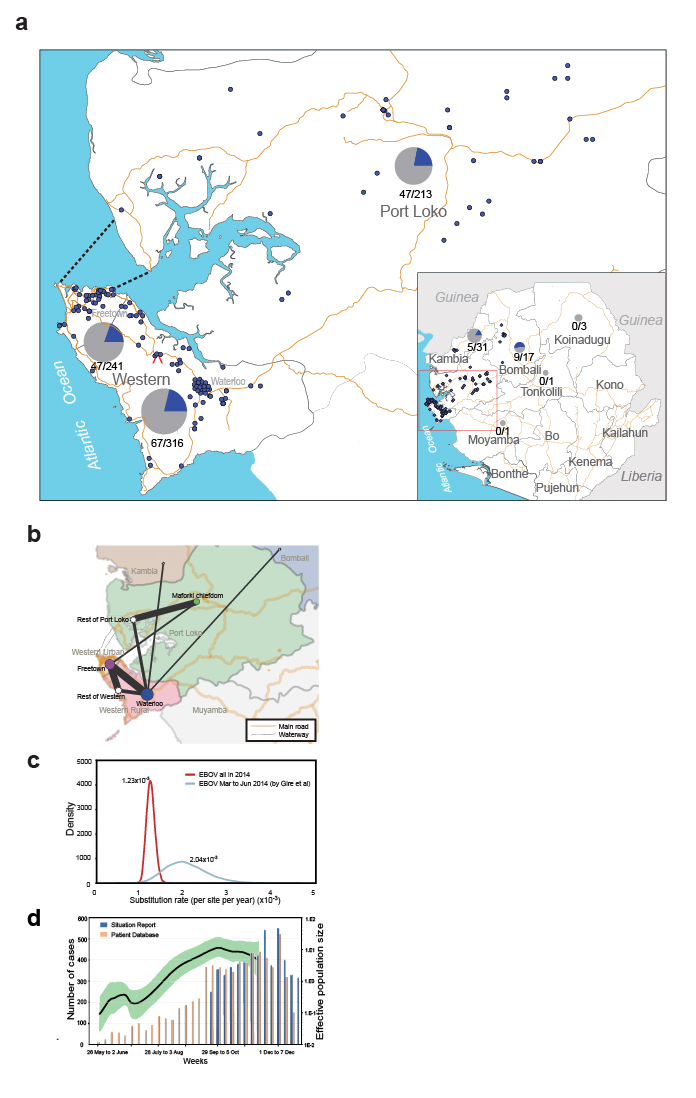
Figure 1. Geographic distribution and evolutionary dynamics of 2014 EBOV. a, geographical distribution of 823 EBOV samples and 175 newly sequenced genomes. b, the phylogeographic linkage of 2014 EBOV in Sierra Leone. c, nucleotide substitution rate of 2014 EBOV. d, growth of the effective population size of 2014 EBOV.
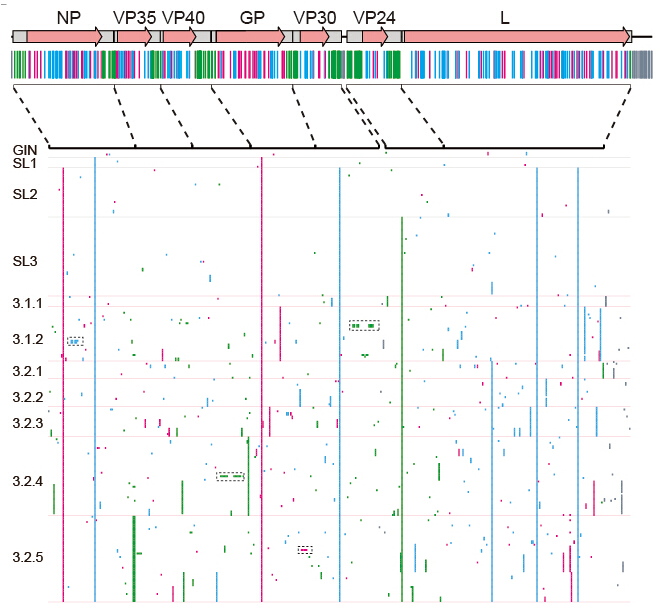
Figure 2, Genomic variations of the 2014 EBOV
Contact:
Prof. George Fu Gao
CAS Key Laboratory of Pathogenic Microbiology & Immunology,Institute of Microbiology,Chinese Academy of Sciences,100101,Beijing, China
E-mail: gaof@im.ac.cn
Dr. LIU Di
Network Information Center, Institute of Microbiology,Chinese Academy of Sciences,100101,Beijing, China
Email: liud@im.ac.cn
