It is well established that regulatory T cells (Treg cells) play a pivotal role in maintaining immune tolerance and thus in preventing autoimmunity and chronic inflammation. These cells can be subdivided into “central” Treg cells population and “effector” Treg cells populations based on function.
Such effector Treg cells make up a minor fraction of them in the circulation and secondary lymphoid organs. They are referred to as “activated” Treg cells in some studies because they share phenotypic features with activated conventional ones which exhibit enhanced regulatory function and migration through non-lymphoid tissues.
Importantly, Treg cells comprising the effector subpopulation defined above are thought to have encountered antigens and have undergone TCR stimulation more recently than central Treg cells.
This coincides with the fact that TCR signaling in differentiated Treg cells is essential to Treg cell homeostasis, suppressor function and signature gene expression, especially to those signature genes that produce Treg cell effector molecules.
It should be noted, however, that the underlying molecular mechanisms downstream of TCR signaling that account for the above Treg cell characteristics are still poorly understood.
One important way in which effector Treg cell differentiation may be regulated is via the activity of E-proteins, a family of transcriptional activators or repressors that bind to E-box sites at transcriptional sites and thus determine gene expression.
Previous studies have shown that down-regulation of E-protein activity by TCR signaling accompanies the development of Foxp3+ regulatory T cells (Ping Gao et. al, JEM, 2014, 211(13):2651-68). Nevertheless, whether E-protein regulates matured Treg cells activation or whether E-protein directly regulates certain activated Treg signature genes expression has never been demonstrated.
Prof. ZHANG Fuping’s group at Institute of Microbiology, Chinese Academy of Sciences (IMCAS) published their research progress entitled “E-protein regulatory network links TCR signaling to effector Treg cell differentiation” on PNAS.
This work reveals that E-protein links TCR signaling to the differentiation of effector Treg cells and elucidates the molecular mechanism by which E-protein regulates the differentiation of effector Treg cells.
They demonstrate that Treg cells lacking E-protein undergo further differentiation into effector cells that exhibit high expression of effector Treg signature genes, including IRF4, ICOS, CD103, KLRG-1 and RORγt.
E-protein deficient Treg cells displayed increased stability and enhanced suppressive capacity. Transcriptome and ChIP-seq analyses revealed that E-protein directly regulates a large proportion of the genes that are specific to effector Treg cells activation.
Importantly, most of the up-regulated genes in E-protein-deficient Treg cells are also TCR-dependent; this indicates that E-proteins comprise a critical gene regulatory network that links TCR signaling to the control of effector Treg cell differentiation and function.
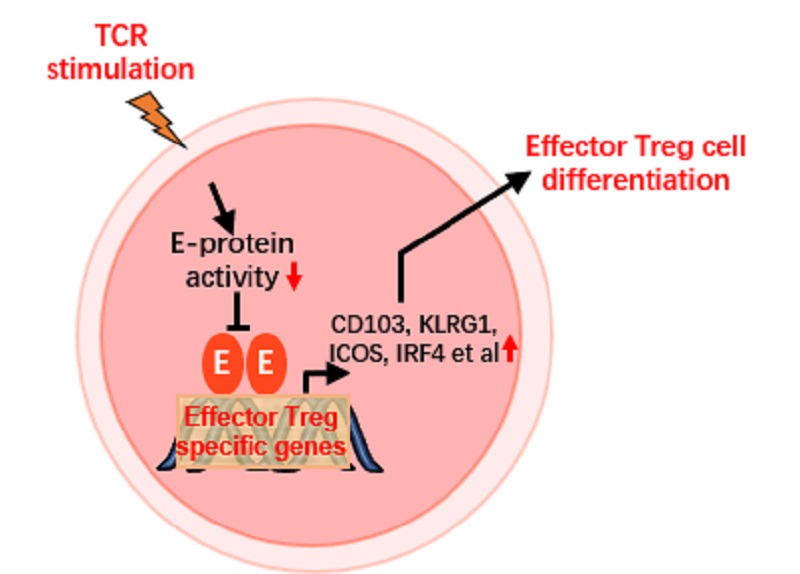
Figure 1. E-protein regulatory network links TCR signaling to effector Treg cell differentiation (Image by Prof. ZHANG Fuping’s goup)
This study reveals that E-protein as a target of TCR signaling directly mediates the regulation of effector Treg cell differentiation, filling the gap of the regulatory network of "TCR signal-effector Treg cells".
Given the central role of regulatory T cells in maintaining autoimmune tolerance, this study will provide new opportunities for regulatory T cells to be used in the treatment of cancer, autoimmune diseases, chronic infections, transplant tolerance and many other aspects.
Dr. HAN Xiaojuan at IMCAS is the first author of the paper, and Prof. ZHANG Fuping, Institute of Microbiology, Chinese Academy of Sciences is the corresponding author of the paper.
This study was supported by the Priority Research Program of the Chinese Academy of Sciences, the National Natural Science Foundation of China, and the National Science and Technology Major Project.