Arenaviruses are a group of enveloped and segmented negative-sense RNA viruses (sNSVs), comprising the family Arenaviridae. All human-infecting arenaviruses belong to the genus mammarenavirus, exemplified by Lassa virus (LASV). LASV is endemic in west Africa which causes approximately 100,000 to 300,000 human infections annually, with a case fatality rate as high as 20%. Unfortunately, there are no specific drugs or vaccines available for most arenavirus infections, posing great challenges for the treatments of related diseases.
Arenaviruses encode their own RNA-dependent RNA polymerase (RdRp), namely the L protein, for replication and transcription of the viral genome, and this molecule is quite conserved among different viral species, which indicates a very important antiviral therapeutic target. Besides, previous studies have revealed that the Z protein could interact with the polymerase to inhibit RNA synthesis at the late stage of viral replication, thus initiate the assembly of progeny virions. Therefore, elucidating the working mechanism of arenavirus polymerase and the regulatory mechanism of Z protein for the polymerase activity is of great significance for developing specific and broad-spectrum antiviral drugs.
In 2020, researchers of Shi lab at the Institute of Microbiology, Chinese Academy of Sciences (CAS), reported the first near-atomic resolution structure of arenavirus polymerase, as well as its complex with viral RNA (vRNA) promoter1. In these structures, they identified a conserved 3’-RNA binding site in different sNSV polymerases. This site would thus be a promising candidate target for developing broad-spectrum antiviral drugs. Besides, they also found the 5’-vRNA regulates the switch between transcription and replication, and the self-dimerization is crucial for the activity of the polymerase1. These findings constitute a basic framework for understanding the mechanism of arenavirus replication, but yet many further questions remain.
Recently, they also investigated the mechanism of arenavirus matrix protein Z regulating the polymerase activity. In this study, they determined the structures of LASV and MACV L proteins in complex with their cognate Z proteins, and the ternary complexes of L-Z-vRNA. These structures reveal that the Z protein binds to the periphery of palm domain of RdRp, far away from the binding site of vRNA. Therefore, the binding of Z protein does not prevent vRNA recruitment by the L polymerase. Despite no catalytic residues being directly engaged by Z protein, the distal ends of two catalytic motifs, D and E, are involved in interactions with Z protein outside the catalytic center. Moreover, these binding motifs are located at the interface between multiple domains. The binding of Z protein would thus restrict the conformational changes of key catalytic motifs required for catalysis, resulting in inactivation of the polymerase2.
Looking into the L-Z contacting interface, they also found that Z protein binds to the L protein through its central domain, in which a highly conserved hydrophobic loop dominates the interaction with the L polymerase. And because of the conservation of the dominant binding motif, they observed that LASV and MACV Z proteins could cross-inhibit the activity of heterologous L polymerases. This observation is quite remarkable. It indicates the regulatory effect of Z protein on polymerase might be a universal mechanism among all arenaviruses, and also suggests the possibility of cross-inhibition between different viruses2. These findings again provide a new strategy for developing broad-spectrum antiviral drugs against different arenaviruses.
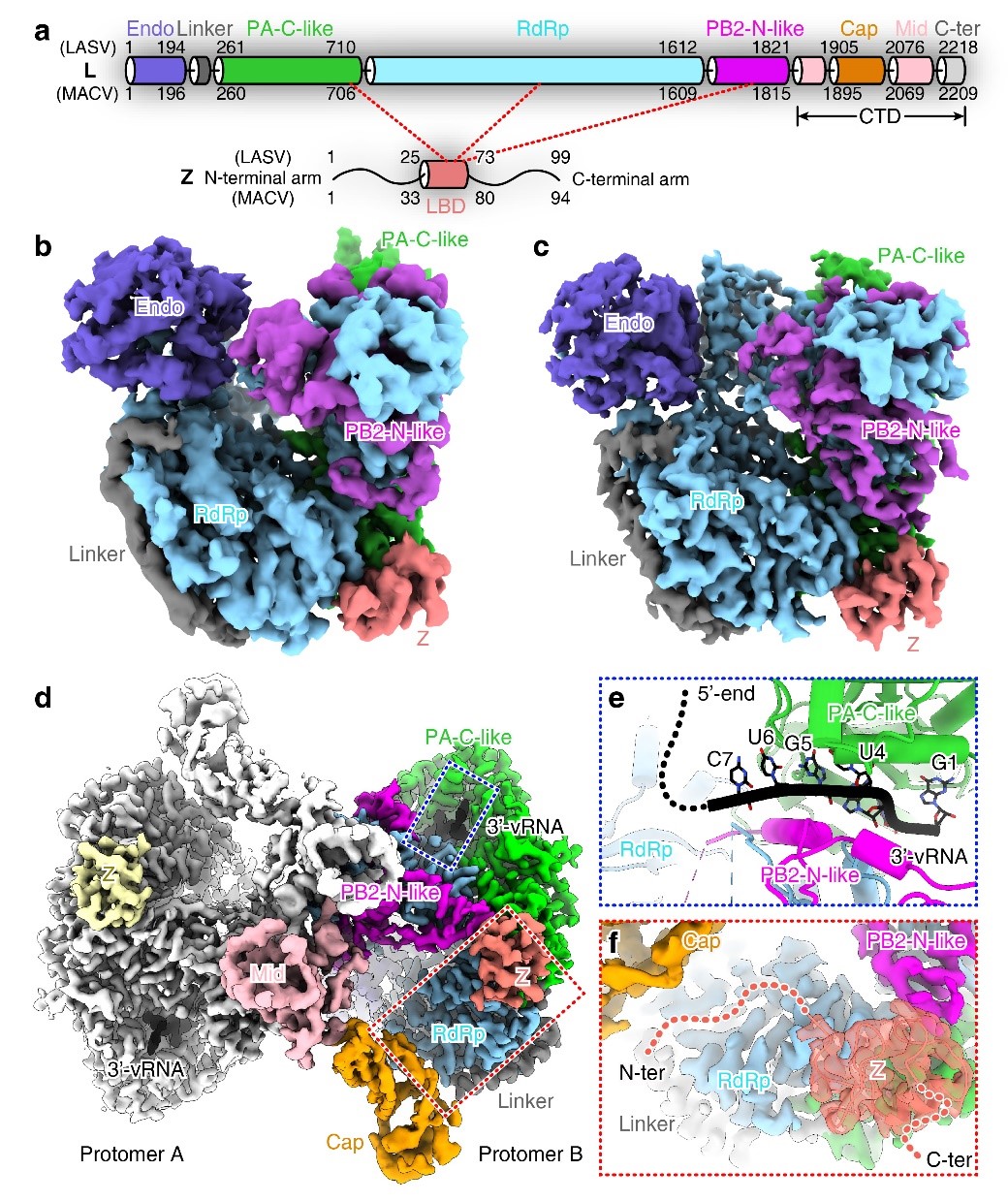
(a) Schematic diagram of domain structures for L and Z proteins. (b-c) Cryo-EM density maps of LASV (b) and MACV (c) L-Z monomeric complexes. (d) The composite map of dimeric complex of MACV L bound to Z and vRNA. (e) Close-up view of the 3'-vRNA binding site. (f) Zoom-in view of the Z protein binding site in the MACV L-Z-vRNA dimeric complex. (Image by Prof. SHI’s group)
References:
2. Xu X. et al. Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.Nat. Microbiol.(2021). DOI: 10.1038/s41564-021-00916-w.