Non-segmented negative-strand RNA viruses (nsNSVs) including Ebola virus (EBOV), can cause respiratory infections, hemorrhagic fever and encephalitis in humans and animals, are considered a substantial health and economic burden worldwide. Although two therapeutic antibodies have been approved to treat Zaire EBOV, effective vaccines and broad-spectrum antiviral drugs are yet lacking.
Replication and transcription of the viral genome are executed by the large L polymerase which is a promising target. During RNA synthesis, the polymerase should bind with genomic RNA and recognize regulatory elements to perform the replication and transcription processes. However, no RNA binding has been observed in the available structures of nsNSV RNA polymerase complexes, and how these nsNSV polymerases recognize the terminal promoter elements remains unknown.
Now, a joint team led by Dr. SHI Yi and Dr. George Fu Gao from the Institute of Microbiology of Chinese Academy of Sciences, have recently investigated the molecular mechanism of de novo replication by the Ebola virus polymerase, which was recently published in Nature. In this study, they demonstrated that the EBOV L-VP35 polymerase complex possesses the de novo replication activity in a leader-sequence-specific manner, and then determined the structure of L-VP35 in complex with the 18-mer leader RNA to a resolution of 3.0 ? using cryo-EM technology (Figure 1).
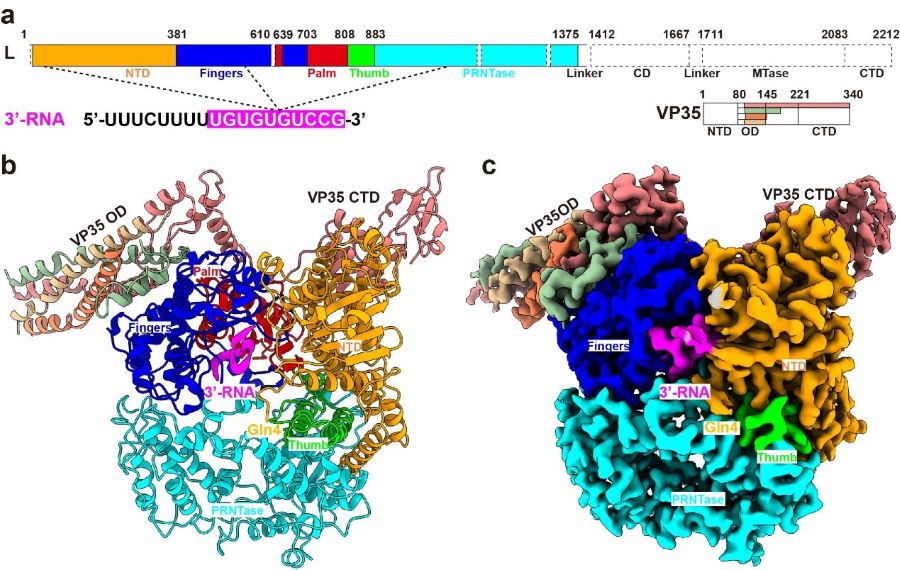
Figure 1 Overall structure of EBOV L-VP35-RNA complex (Image by SHI Yi)
In the structure, the 3’ leader RNA binds in a distinctive tortuous conformation in the template entry channel of the L protein, and many intramolecular/intermolecular interactions are essential for maintaining the conformation. Notably, G7 nucleotide flips to the opposite direction, and is stabilized by three hydrogen bonds (Figure 2). When the G7 was substituted with other nucleotides, the de novo replication activity was completely abolished in vitro. They further evaluated that the distinctive bend conformation of RNA is more required for the de novo replication activity rather than the elongation activity.
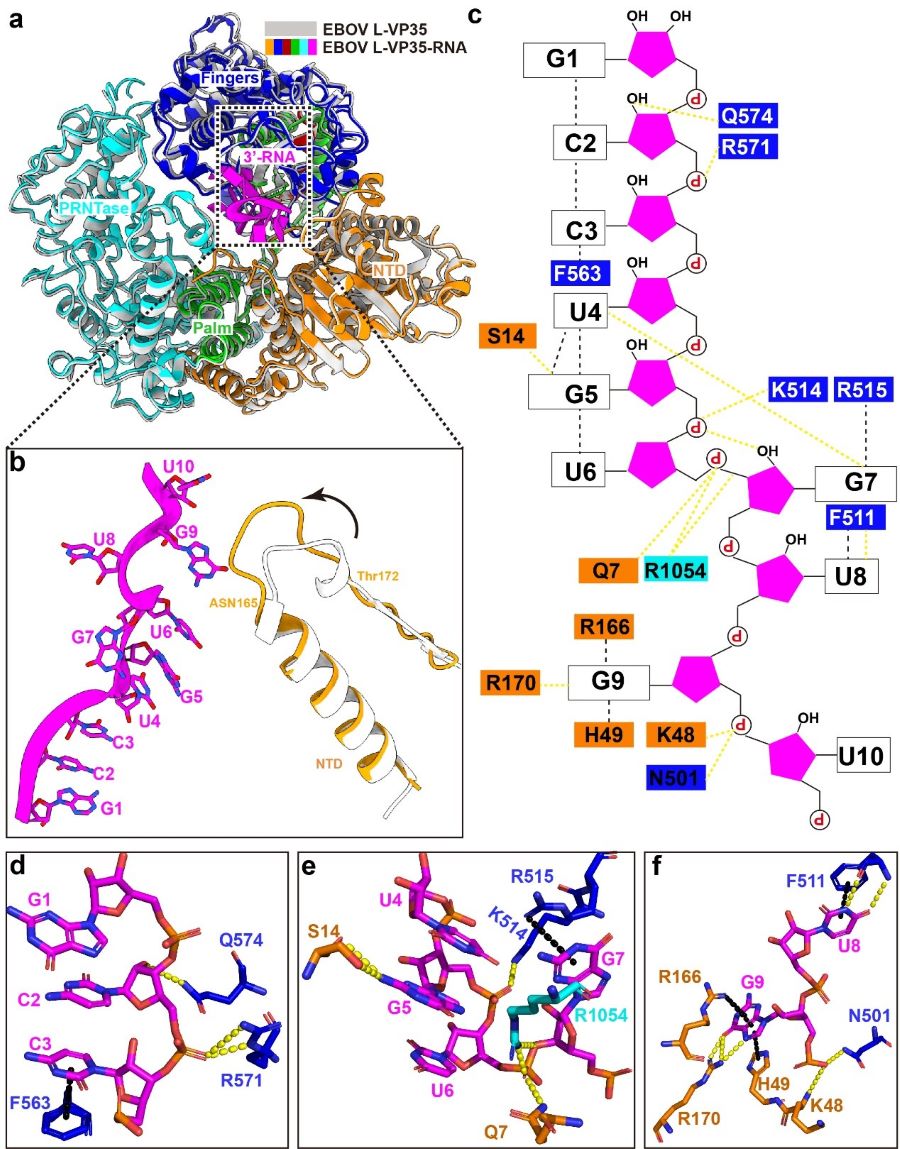
Figure 2 Molecular interaction between the EBOV polymerase and the leader RNA (Image by SHI Yi)
In contrast to the segmented negative-strand viruses (sNSVs) including influenza viruses and bunyaviruses, the EBOV RNA promoter does not bind in a positively charged groove at the peripheral surface of polymerase protein, and instead, the 3’ RNA promoter is directly accommodated in the template entry channel with a distinctive bend conformation, proposed that the polymerases of nsNSVs including EBOV have evolved a different replication initiation mechanism to adapt their RNP structure (Figure 3).
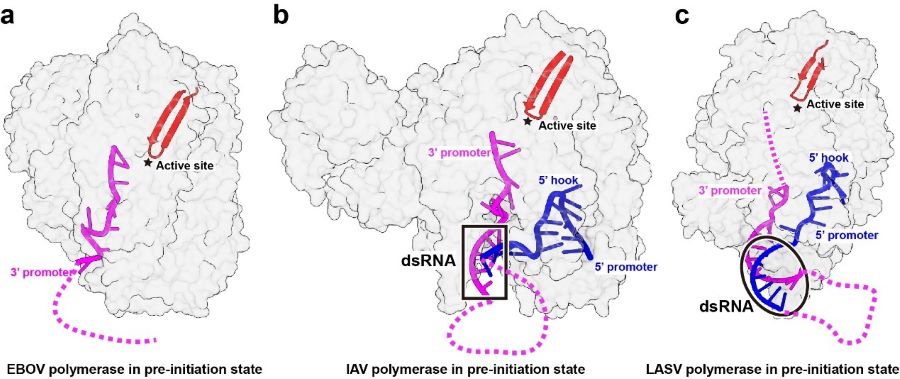
Figure 3 Structural comparison of different viral polymerases in the pre-initiation state (Image by SHI Yi)
This study provided the first structural snapshot of EBOV L polymerase recognizing the genome RNA and revealed a distinctive mechanism of replication initiation for nsNSVs. will greatly enhance our mechanistic understanding of RNA synthesis by nsNSV RNA polymerases, and open avenues to the development of new antiviral agents that target the polymerases of nsNSVs.
Contact
SHI Yi
Institute of Microbiology of Chinese
Phone: +86-10-64806050
E-mail: shiyi@im.ac.cn