For decades, the ribosome has been known as one of the most complicated, orchestrated, and highly conserved molecular machine for translating various mRNAs into proteins, being essential for growth and proliferation in cells of all organisms. Generation of the ribosome is a high energy-consuming process, as it requires all three RNA polymerases and more than 150 non-ribosomal factors. Generally, ribosome-mediated translation and ribosome biogenesis are considered housekeeping processes, but new research from Institute of Microbiology, Chinese Academy of Sciences (IMCAS), has revealed ribosomes are not simple machines but also can function as translational regulatory elements in controlling activation of regulatory T cells (Tregs).
Tregs are immune cells with potent suppressive activity, working to dampen unwanted autoimmune response that attacks normal tissue and cells. Tregs can be divided into central Tregs (cTregs) and effector Tregs (eTregs) according to their activation status. The transition from cTregs to eTregs relies on TCR stimulation signals delivered by antigen presenting cells (APCs). Recently, global transcriptomic and proteomic analyses of Tregs have illustrated remarkable discordance between mRNA and protein datasets, highlighting the importance of post-transcriptional regulation. Thus far, the translational regulation mechanism in determining the activation status of Tregs is poorly understood.
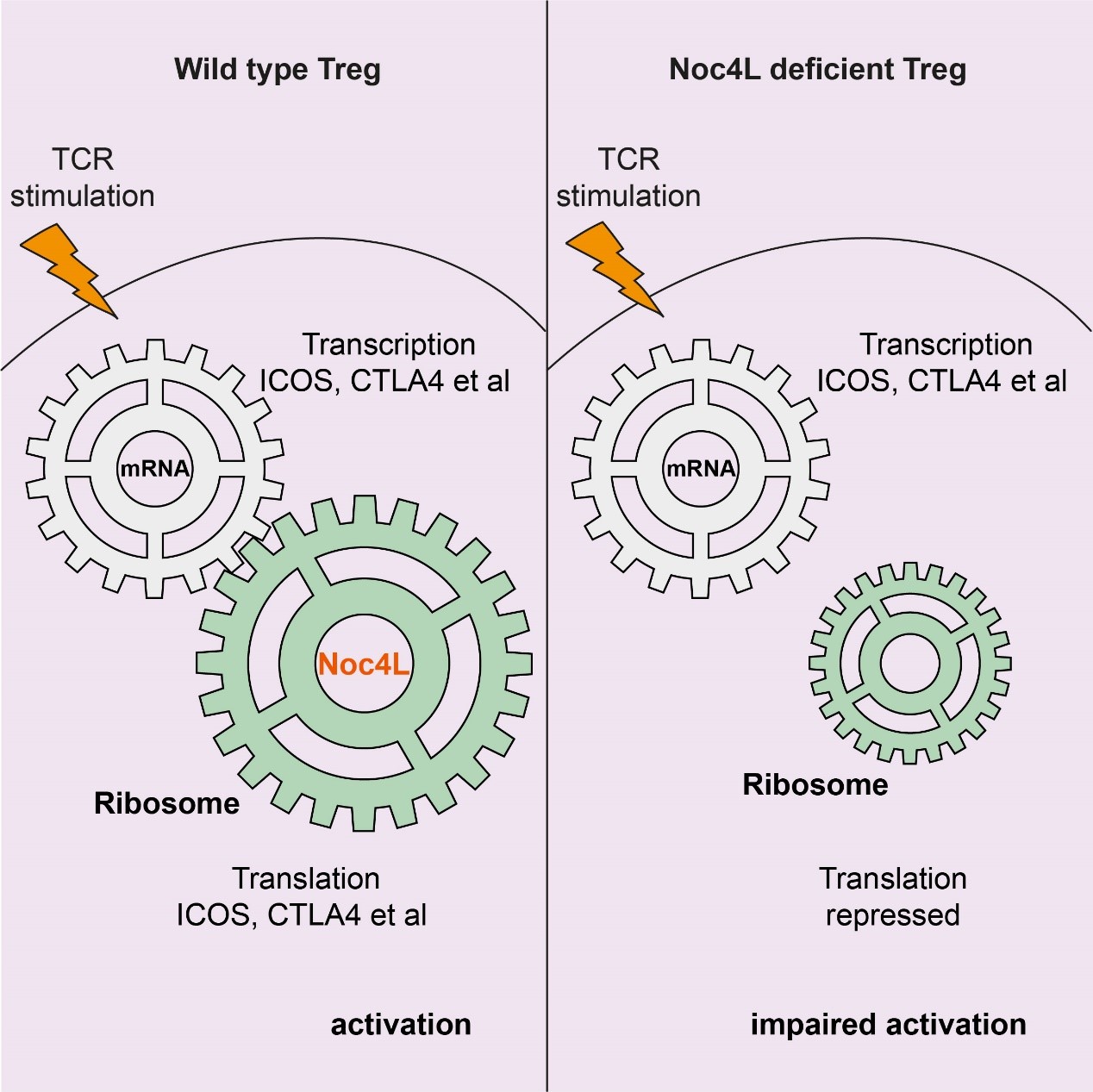
Published this week in Cell Reports, researchers from IMCAS focused on a protein called Noc4L, which is essential for mammalian ribosome biogenesis. Noc4L is not uniformly expressed in all tissues but is preferentially expressed in lymphoid organs, and human Noc4L is located in multiple sclerosis (MS) susceptibility locus at 12q24.33. But the functional role of Noc4L in ribosome biogenesis in Tregs is unknown. Xueping Zhu et al. found Treg specific Noc4L knockout mice developed a lethal autoimmune phenotype, resembling Treg deficient “scurfy” mice. Unexpectedly, Noc4L knockout did not globally affect the overall protein translation in Tregs, but rather selectively affected the translation of mRNAs (Icos, Ctla4, Cd69, etc.) related to Treg activation. Subsequent experiments revealed the limiting of the ribosome concentration is the main cause of the selective translation failure in Noc4L deficient T cells. These findings represent a significant step forward in the understanding of the immune system, and Noc4L-mediated ribosome biogenesis factors might serve as new targets for clinical manipulation of Tregs to control autoimmune diseases.
Journal Reference:
Noc4L-Mediated Ribosome Biogenesis Controls Activation of Regulatory and Conventional T Cells. Zhu X, Zhang W, Guo J, Zhang X, Li L, Wang T, Yan J, Zhang F, Hou B, Gao N, Gao GF, Zhou X. Cell Rep. 2019 Apr 23;27(4):1205-1220.e4. doi: 10.1016/j.celrep.2019.03.083.
Paper link: https://www.cell.com/cell-reports/fulltext/S2211-1247(19)30422-X
Tregs: Lost in Translation
Contact:
Dr.ZHOU Xuyu
CAS Key Laboratory of Pathogenic Microbiology and Immunology,Institute Of Microbiology,Chinese Academy of Sciences,100101,Beijing, China
E-mail: zhouxy@im.ac.cn