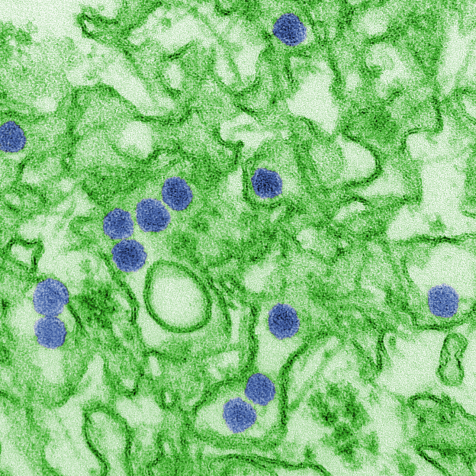
This is a digitally-colorized transmission electron micrograph (TEM) of Zika virus, which is a member of the family Flaviviridae. Virus particles, here colored blue, are 40 nm in diameter, with an outer envelope, and an inner dense core. Credit: CDC/ Cynthia Goldsmith, Wikipedia
Zika virus, the name sounds more familiar these days than many famous celebrities. With Singapore reporting its first case of Zika virus last month, CDC and WHO declaring ZIKA as global pandemic and with health advisory officially advising that Rio Olympics must be postponed, ZIKA virus has certainly caught the attention of one and all.
Though it is the frontline news these days, Zika virus was first discovered in 1947 and is named after the Zika Forest in Uganda. First human cases of Zika were detected and reported since then, and outbreaks of Zika have been reported in tropical Africa, Southeast Asia, and the Pacific Islands. Before 2007, at least 14 cases of Zika had been documented, although other cases were likely to have occurred unreported. Since the symptoms of Zika are similar to many other diseases, many cases may not have been recognized then.
In May 2015, the Pan American Health Organization (PAHO) issued an alert regarding the first confirmed Zika virus infection in Brazil. On February 1, 2016, the World Health Organization (WHO) declared Zika virus a Public Health Emergency of International Concern (PHEIC). Local transmission has been reported in many other countries and territories. Zika virus will likely continue to spread to new areas.
Zika virus (ZIKV), a mosquito-borne flavivirus belongs to a family of viruses that have single-stranded positive sense RNA. In simple terms, these viruses have only single-stranded genetic material and direction of RNA is 5′ to 3′ allowing direct translation into the desired viral proteins.
The recently published cryoelectron microscopy (cryo-EM) structures of ZIKV reveal that ZIKV displays a similar structure to other known flaviviruses. For flavivirus, mature virus particles contain 180 copies of the envelope (E) protein and membrane (M) protein on the envelope and display an icosahedral arrangement in which 90 E dimers completely cover the viral surface
The flavivirus envelope (E) glycoprotein is responsible for virus entry and represents a major target of neutralizing antibodies for other flaviviruses.
Scientist at Institute of Microbiology, Chinese Academy of Sciences have now solved structures of ZIKV E protein at 2.0 A° resolution using X-ray crystallography. The team led by Prof. George F. Gao was also successful in solving the structure of E protein in complex with a flavivirus broadly neutralizing murine antibody 2A10G6 at 3.0A°.
Structural comparison of ZIKV-E with other members of family flaviviridae shows that although structure of the protein is very much similar, it contains a unique, positively charged patch which may influence host attachment. The ZIKV-E-2A10G6 complex structure reveals antibody recognition of a highly conserved fusion loop. 2A10G6 binds to ZIKV-E with high affinity invitro and neutralizes currently circulating ZIKV strains in vitro and in mice. Therefore, the E protein fusion loop epitope represents a potential candidate for therapeutic antibodies against ZIKV (Biotechin.Asia).
https://biotechin.asia/2016/06/24/structural-biologists-elucidate-structure-of-zika-virus-envelope-protein/
Contact:
Prof. GAO Fu
CAS Key Laboratory of Pathogenic Microbiology and Immunology,Institute of Microbiology,Chinese Academy of Sciences,100101,Beijing, China
E-mail: gaof@im.ac.cn