Scientists have elucidated the structure of a key protein that helps the Zika virus to infect cells. GAO Fu (George F. Gao) and SHI Yi from Institute of Microbiology, Chinese Academy of Sciences in Beijing and their colleagues crystallized the NS1 protein from the Zika virus involved in the 2015 outbreak in Brazil.
Zika virus (ZIKV), a Flaviviridae family member, transmitted to humans by mosquitoes of the genus Aedes, was first isolated in Africa in 1947 and found to commonly circulate in the tropical regions in Africa and Asia. Following a large Zika outbreak in French Polynesia from 2013 to 2014, the virus was able to emerge into new territories of the American continent and has been found to be circulating in 26 countries and territories in South America and the Caribbean.
Prior to this outbreak, research into ZIKV pathogenesis was largely neglected, as infected individuals can often be asymptomatic or have mild symptoms. There is now growing evidence that ZIKV infections may be linked to fetal and newborn microcephaly and serious neurological complications, such as Guillain-Barre syndrome (GBS).
ZIKV has been detected in the amniotic fluid of pregnant women whose fetuses had microcephaly syndrome, and also in microcephalic fetal brain tissues. Moreover, ZIKV infects human cortical neural progenitor cells and attenuates their growth. To date, no clinically-approved vaccines or therapeutics are available to prevent and control ZIKV infection.
Under this urgent situation, great efforts are called for in order to develop novel vaccines and antiviral therapeutics, for which a comprehensive understanding of the pathogenesis and molecular basis of ZIKV infection is required.
As a member of Flaviviridae family, ZIKV has a single positive sense RNA genome, initially translated as a single polyprotein which is then cleaved post-translationally into three structural proteins (C, PrM or M, E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5).
The glycosylated NS1, which associates with lipids, forms a homodimer inside the cells and is necessary for viral replication and late in infection. NS1 is also secreted into the extracellular space as a hexameric lipoprotein particle, which is involved in immune evasion and pathogenesis by interacting with components from both innate and adaptive immune systems, as well as other host factors. NS1 is the major antigenic marker for viral infection, and has been suggested as a biomarker for early detection of dengue virus infection combined with other markers.
Dr. GAO and SHI’s research groups report that the ZIKV NS1 protein has structural similarities to those found in closely related viruses such as the dengue and West Nile viruses. But NS1 has some key differences from these related proteins — most notably, a different pattern of electric-charge distribution in a region that interacts with host cells.
The DENV1 and DENV2 NS1 structures both display a positively charged surface in the central region of loop surfaces, whereas the WNV NS1 structure has a negatively charged central region in the loop surface. For ZIKV, the loop surface exhibits a composite platform containing both a positively and negatively charged central region and a negative charge observed towards the two distal ends.
Their article entitled “Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses” was published in Nature Structural & Molecular Biology (http://www.nature.com/nsmb/journal/v23/n5/full/nsmb.3213.html) on April 18 2016. It is selected by Nature in Research Highlights ( http://www.nature.com/nature/journal/v532/n7599/full/532285a.html ).
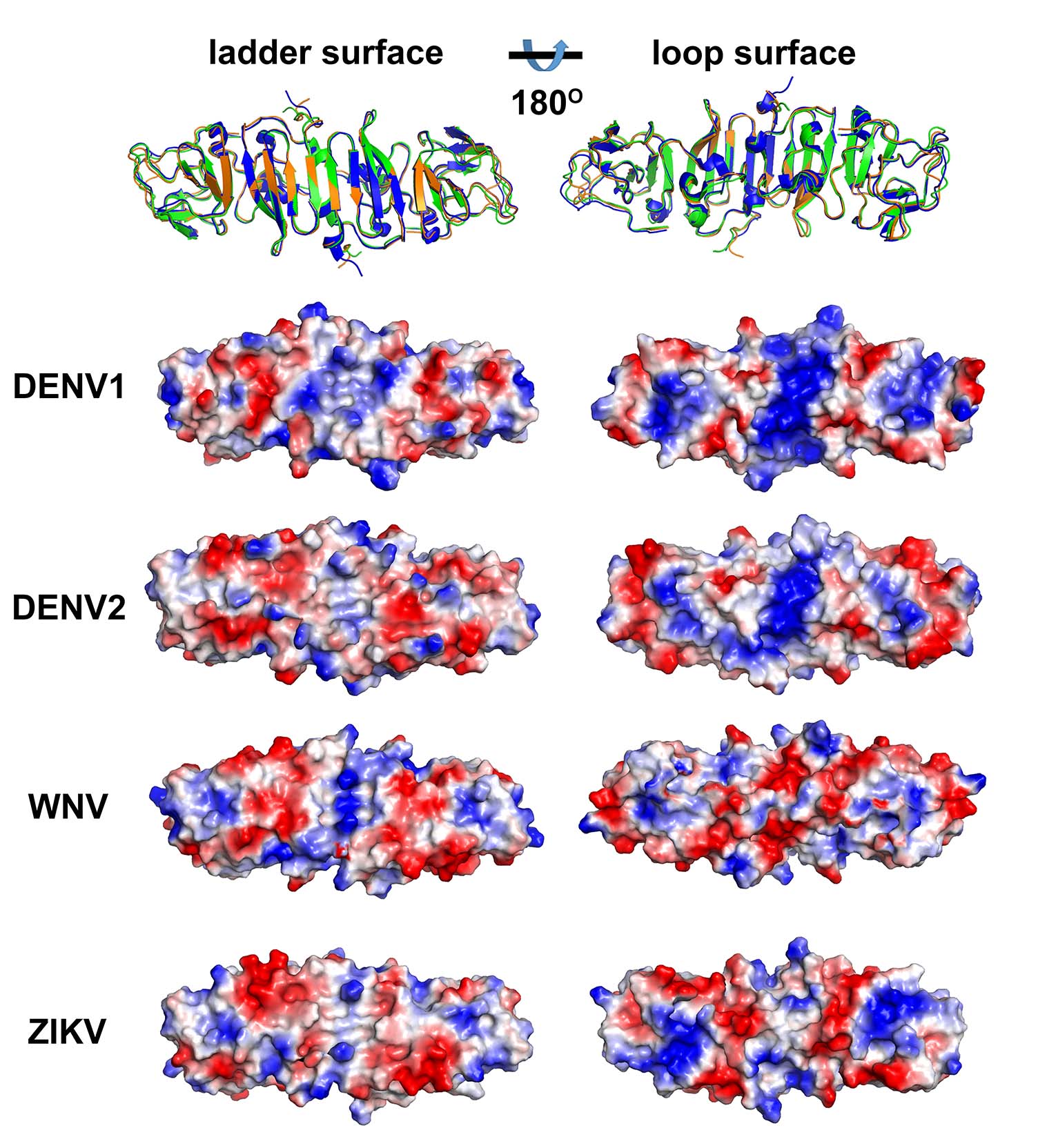
Comparison of ZIKV NS1 with other known NS1 structures (Image by Prof. GAO’s group)
Key words: Zika virus, crystal structure, hexameric lipoprotein
Contact:
Prof. GAO Fu
CAS Key Laboratory of Pathogenic Microbiology & Immunology
E-mail: gaof@im.ac.cn