Ebola virus can cause a rapidly-lethal hemorrhagic fever in humans and primate animals, and currently no clinically approved antiviral therapeutics is available. Since 1976, Ebola virus has emerged multiple times, with case fatality rates reaching ~90%. During 2013-2015, a historically-unprecedented Ebola epidemic has swept through the West Africa, causing more than 28,000 human infections and over 11,000 related deaths as of January 10th, 2016.
Under the urgent circumstances, it is called for great efforts to develop the vaccines and antiviral therapeutics, which requires comprehensive and in-depth understanding of this devastating virus.
In September 2014, China sent its first aid group to Sierra Leone to help control Ebola outbreak. George F. Gao was the deputy director of the aid group and he also took charge of communication and coordination with international organizations. During his stay in Sierra Leone, he wrote a paper entitled “On the Ground in Sierra Leone” in Science, describing the real working life there. Subsequently, he and his colleagues analyzed the genomes of Ebola viruses in Sierra Leone, and published their findings in Nature.
After he returned to China, he and his group still focused on Ebola virus basic researches. On January 14 2016, an article entitled “Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1” was published in Cell by George F. Gao’s group from Institute of Microbiology, Chinese Academy of Sciences. WANG Han, SHI Yi, SONG Jian and QI Jianxun are the co-first authors, and George F. Gao is the corresponding author.
They have elucidated the molecular basis of how the primed Ebola viral glycoprotein binds to its endosomal receptor NPC1, and proposed a fifth membrane fusion trigger mechanism of enveloped viruses, which differs from the previous four membrane fusion mechanisms. Their findings provided a structural insight into Ebola virus entry in the late endosome and the guidance for design of therapeutic inhibitors.
Regarding the infection by Ebola virus, the virus entry process includes two key steps, virus attachment on the cell membrane and virus membrane fusion in the endosome.
George F. Gao’s group has studied the host cell attachment factor, T cell immunoglobulin and mucin domain-containing (TIM) proteins, and elucidated the molecular basis of interaction between the TIM proteins and their ligand phosphatidylserine (PS), shedding light on the role of TIM proteins in enhancing Ebola virus infection. This finding was published in Chinese Journal "Chinese Science bulletin" as a cover story.
Following binding to the cell surface, Ebola viruses are internalized by a macropinocytosis-like process and subsequently trafficked through early and late endosomes. The trimeric glycoprotein (GP) spike on the envelope of Ebola viruses mediates all stages of virus entry, including membrane fusion.
Like all the other Class I viral fusion proteins, the EBOV GP is synthesized as a single polypeptide that is subsequently cleaved by furin-like proteases into GP1 and GP2 subunits, which remain together through an inter-subunit disulfide bond and non-covalent interactions.
However, in contrast to other Class I viral fusion proteins, the simple furin cleavage event is not sufficient to prime EBOV GP. After entering into the cell, the virus is eventually trafficked to late endosomes, where GP is further primed to remove some “cap” components, thereby triggering the induction of the crucial membrane fusion event which leads to viral penetration.
The functional EBOV GP priming is mediated by the cysteine proteases cathepsin B and cathepsin L, resulting in a primed form of GP (GPcl). Unlike the full-length GP, the primed GPcl could bind to an endosomal membrane protein Niemann-Pick C1 (NPC1), which is an indispensable host entry factor for EBOV infection.
Here, for the first time, they have determined the crystal structures of free NPC1-C and its complex with GPcl. The results revealed that NPC1-C displays a helical structure core surrounded by several β-strands, and contains two extended loops protruding outwards for ligand interactions.
Further GPcl/NPC1-C complex structure indeed shows that NPC1-C utilizes the two protruding loops to engage a hydrophobic cavity on the head of GPcl.
Despite a low affinity between NPC1-C and GPcl as revealed by the biophysical analyses, upon enzymatic cleavage and NPC1-C binding, several conformational changes are shown to be induced in GPcl, including the uplift of the short 310 helix in the β3-α1 loop likely helps to release the N-terminal portion of the internal fusion loop (IFL), thereby triggering the membrane fusion. The mutagenesis work in NPC1-C further confirmed that the protruding loop 2 plays a more important role in the binding of NPC1-C to GPcl.
The results for the structural basis of primed EBOV GP bound to its endosome-residing receptor NPC1 expands understanding in the fusion trigger mechanism of enveloped viruses, and provides valuable information for the design of therapeutics against Ebola virus infections.
The model of Ebola virus invasion(Image by GAO)
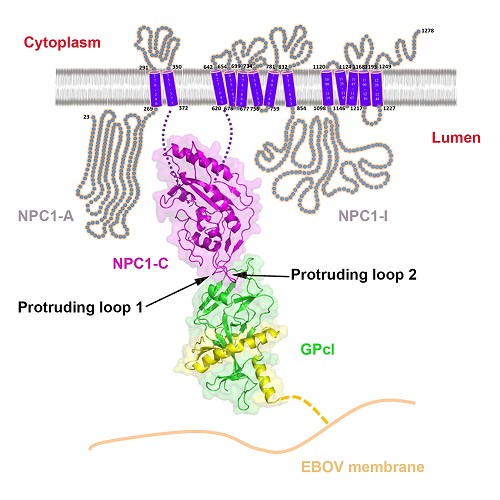
The details of interaction between Ebola virus and its endosomal receptor(Image by GAO)