Hemerythrin-like proteins are non-heme, di-iron and O2-binding proteins that are ubiquitous from bacteria to mammals and have been shown to function in oxygen storage and transport. Although hundreds of hemerythrin-like proteins have been predicted in bacteria, studies on the biological functions of hemerythrin-like proteins are scarce. Recently, a study showed that the multi-domain protein VcBhr-DGC (with a hemerythrin domain and a diguanylate cyclase GGDEF domain) functions as a regulatory oxygen sensor for switching between reducing or anaerobic environments in Vibrio cholerae. This is the first one to demonstrate a regulatory function of the hemerythrin domain and hints that hemerythrin-like proteins might have other unannotated functions. As a follow-up of that study, members of the lab of Dr. Mi from the Institute of Microbiology have analyzed the function and relationship between multiple hemerythrin-like proteins withinMycobacterium smegmatis, the model for Mycobacterium tuberculosis.
M. smegmatis contains three hemerythrin-like proteins, MSMEG_3312, MSMEG_2415 and MSMEG_6212. Using knock-out, complementation and overexpression strains of these genes, researchers from the Mi lab found that MSMEG_2415 was involved in the response to H2O2. In addition, using promoter regulation experiments, it was shown that MSMEG_2415 regulates sigF expression via promoter Prbsw and is essential for inhibition of the SigF-mediated H2O2 response (Figure 1). To our knowledge, MSMEG_2415 is the first characterized hemerythrin-like protein in mycobacteria.
Next, the genes encoding the two other hemerythrin-like proteins in M. smegmatis were investigated in a similar manner. Experiments showed that MSMEG_6212 and MSMEG_3312 both modulate erythromycin susceptibility and the msmeg_3312 and msmeg_6212 double-knockout strain, mc2155:Δ3312-6212, has a similar resistance as the individual single-knockout strains mc2155:Δ3312 and mc2155:Δ6212. Moreover, MSMEG_3312 and MSMEG_6212 were found to inhibit MtrA-mediated erythromycin resistance, but had no effect on WhiB7-mediated erythromycin resistance. These results are the first to show that MSMEG_3312 and MSMEG_6212 normally lowers the mRNA level of MtrA,thus affecting its regulon, and thereby causing drug resistance (Figure 1).
Interestingly, phylogenetic analysis of bacterial hemerythrin-like proteins showed that the three mycobacterial hemerythrin-like proteins are likely derived from different lineages, as MSMEG_3312, MSMEG_2415 and MSMEG_6212 are present in three different clades (Figure 2). All the proteins in the MSMEG_2415 cluster belong to the Mycobacterium genus, while those in the MSMEG_3312 cluster were derived from Mycobacterium, Rhodococcus and Sciscionellas. A large portion of the hemerythrin-like proteins that were present in the MSMEG_6212 cluster belong to the Streptomyces, Saccharomonospora and Amycolatopsis genera, suggesting that msmeg_6212 has an independent origin. Differences in origins may explain the differences in their physiological functions.
This study is the first to analyze the function and relationship between multiple hemerythrin-like proteins within one organism. The functional and phylogenetic analyses of hemerythrin-like proteins in M. smegmatis has provided insight into the evolutionary selection of antimicrobial resistant traits.

Figure 1 Proposed model for the role of M. smegmatis hemerythrin-like proteins in susceptibility to H2O2 and erythromycin (Image from Dr Mi's lab)
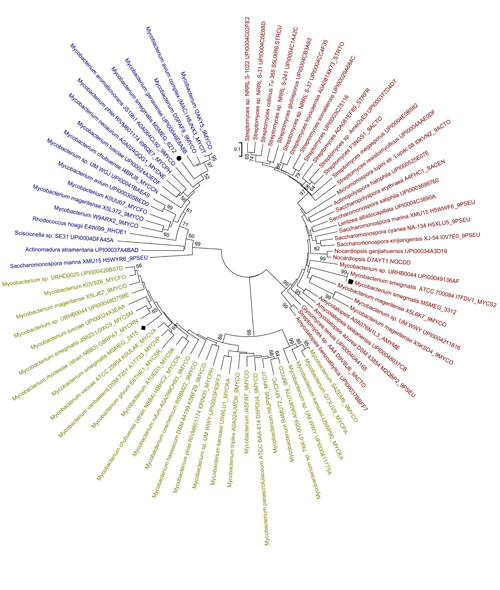
Figure 2:Phylogenetic relationships of mycobacterial hemerythrin-like proteins in bacteria. Squares indicate MSMEG_3312, diamonds indicate MSMEG_2415 and circles indicate MSMEG_6212. (Image from Dr Mi's lab)
E. Key words: hemerythrin-like protein, hydrogen peroxide, erythromycin
F. Funding: These works was supported by the Ministry of Science and Technology of China (2014CB744402 and 2012CB518700), National Natural Science Foundation of China (31270178) and the Key Program of the Chinese Academy of Sciences (KJZD-EW-L02).
G. Article website:
http://journal.frontiersin.org/article/10.3389/fmicb.2014.00800/abstract
http://www.nature.com/articles/srep16130
H.Contact:
Dr. Mi Kaixia
CAS Key Laboratory of Pathogenic Microbiology and Immunology,Institute Of Microbiology,Chinese Academy of Sciences,100101,Beijing, China
E-mail: mik@im.ac.cn