In February of this year, a paper published in Proceedings of the National Academy of Sciences of the United States of America (PNAS) drew worldwide attention, reporting a novel influenza virus genome discovered in several bats from Guatemala. Phylogenetically, this bat-derived viral genome is highly divergent in sequence from other known influenza A viruses, and was thereby designated as a new influenza subtype of H17N10.
Previously identified influenza A viruses includes sixteen (H1-H16) and nine (N1-N9) serotypes for hemagglutinin (HA) and neuraminidase (NA), respectively. The bat-derived influenza virus genome has introduced a seventeenth serotype (H17) in HA and a tenth serotype (N10) in NA. Nothing was known about the capability of H17N10 to cause human infections and its putative pandemic potential.
Prof. GAO Fu (George F. Gao)'s laboratory in Institute of Microbiology, Chinese Academy of Sciences focused on the structural and functional analysis of influenza glycoproteins HA and NA, and studies of the mechanism of influenza cross-species transmission. As a swift response to this new "virus", the neuraminidase N10 was comprehensively studied in his laboratory from a structural and functional perspective.
NA is an important surface glycoprotein of the influenza virus. While HA allows the cell entry of influenza viruses by binding to sialic acid containing receptors, NA helps the release of the newly-generated progeny viruses from infected cells by destroying the receptor. NA is therefore important in the infection, transmission and pathogenesis of influenza.
In this study, Prof. GAO's research group purified the N10 protein from a baculovirus system and performed detailed research on its structure and function. Interestingly, the N10 protein was found to lack canonical NA activity in a fluorescence-based sialidase assay. The crystal structure of N10 was also successfully solved at high resolution, providing an explanation for this lack of canonical activity. As with other NAs, N10 also forms a tetramer in structure with each monomer containing a surface concave that is similar to the catalytic centre of the canonical NAs. Nevertheless, detailed analysis revealed dramatic alterations in the conserved active site residues that are unfavorable for the binding and cleavage of terminally linked sialic acid residues.
These findings imply that H17N10 may have a completely different mechanism for infection of the host cells and could have more restricted host specificity. Consequently, the new bat "virus" is unlikely to transmit to humans in the near future. This study on influenza N10 sheds light on the molecular mechanism of bat influenza virus infection and also provides novel insights into the structure and function of the sialidase superfamily.
These results were published in PNAS in September.
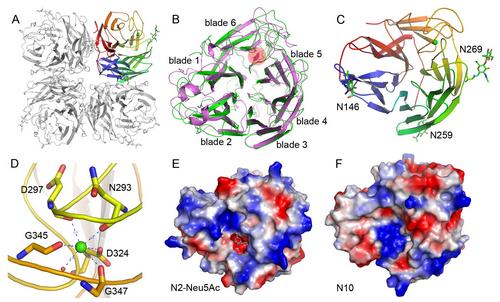
Overall crystal structure of N10 (Image by Prof. GAO’s lab)