Recent study in Institute of Microbiology, Chinese Academy of Sciences (IMCAS) increases understanding about the fungal community structure of Ophiocordyceps sinensis. Fungi other than O. sinensis originating from natural O. sinensis specimens could be one kind of important resources for developing alternative products of natural O. sinensis specimens.
Ophiocordyceps sinensis (syn. Cordyceps sinensis), which is a parasite of caterpillars and is endemic to alpine regions on the Tibetan Plateau, is one of the most valuable medicinal fungi in the world. “Natural O. sinensis specimens” harbor various other fungi. Besides O. sinensis, 10 or more other fungal species have been isolated from natural O. sinensis specimens. Some of these fungi can synthesize chemical components and/or have pharmaceutical value similar to O. sinensis, and fermented mycelial products have been developed and applied in clinical practice. Nevertheless, the mycobiota of natural O. sinensis specimens has not been investigated in detail.
Based on the technique of PCR-single-strand conformation polymorphism (PCR-SSCP), the mycobiota of three different sections (stromata, sclerotia, and mycelial cortices) from natural O. sinensis specimens were investigated using both culture-dependent and -independent methods. For the culture-dependent method, 572 fungal strains were isolated, and 92 putative operational taxonomic units (OTUs) were identified from 226 sequenced strains with the threshold of 97%. For the culture-independent method, 490 fungal clones were identified from about 3000 clones of ITS fragments from the whole-community DNA; based on PCR-SSCP analyses, 266 of these clones were selected to be sequenced, and 118 putative OTUs were detected. The overwhelming majority of isolates / clones and OTUs were detected from mycelial cortices; only a few were detected from stromata and sclerotia. The most common OTUs detected with both methods belonged to Ascomycota; however, only 13 OTUs were detected simultaneously by both methods. Potential novel lineages were detected by each of the two methods.
This research was published in PLoS ONE 5(12): e15570. doi:10.1371/journal.pone. 0015570.
RESEARCHER CONTACT:
LIU Xingzhong, Ph.D., Professor
E-mail: liuxz@im.ac.cn
Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
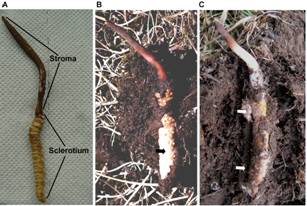
Natural O. sinensis specimens
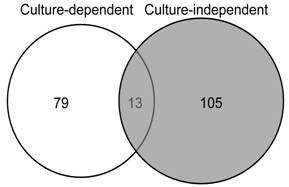
Shared and unique OTUs detected in both investigations