
Principal Investigator,Professor : Bacterial Dormancy and Resistance Mechanism Research Group
1. How bacterial spores exit from their dormancy
2. The molecular basis of bacterial resistance to antibiotics or environmental stress
3. The mechanism of antimicrobial action
Antimicrobial resistance(AMR) is one of the top scientific issues with global concerns, and understanding the mechanisms of bacterial resistance under different physiological conditions is critical for addressing this challenge. My research group is mainly focused on the mechanisms of stress resistance and drug resistance in bacteria under both dormant and vegetatively growing states, aiming to uncover the underlying processes of resistance formation, providing new insights for the development of novel antimicrobial strategies.
1. The molecular basis of bacterial spore germination.
Some bacteria in the orders Bacillales and Clostridiales, can resist antibiotics and sterilization by entering a highly durable spore state. Spores are metabolically inactive and can remain dormant for decades, yet upon exposure to nutrients they rapidly resume vegetative growth and cause food spoilage, food-bore illness, or life-threatening diseases. The exit from dormancy, germination, is a key target in addressing these.
Based on the protein structure modeling predictions and bioinformatics analysis, My group aims to investigate the detailed molecular basis of signal detection and transduction between the spore germination involved key players, via various chemical and biological techniques, providing the scientific evidences to fight against the stress-resistance spores and develop the novel antimicrobial strategies.
2. The mechanism of bacterial stress resistance
In response to the external stress signals, some bacteria are able to survive and adapt to environmental stress by regulating its physiological processes such as cell growth and division. This adaption allows them to escape from the cell damages caused by antibiotics and other physical and chemical factors. In-depth research into the detection of environmental stress signals, and the subsequent signal transduction mechanism, will help uncover the molecular basis of bacterial resistance formation, providing the theoretical support for the development of novel antimicrobial drugs.
3. The mechanism of antimicrobial action
Currently, many natural or synthetic antimicrobial molecules and antimicrobial peptides, exhibit excellent antibacterial activity. However, their specific mechanism of action and drug-targets remain unclear yet. My group aims to identify their precise drug-targets by investigating the antimicrobial mechanisms of these agents, ensuring the screened antimicrobial drugs that effectively inhibit bacteria without damaging human cells as well. This research will provide scientific evidences for clinical applications and the market launch of novel drugs.
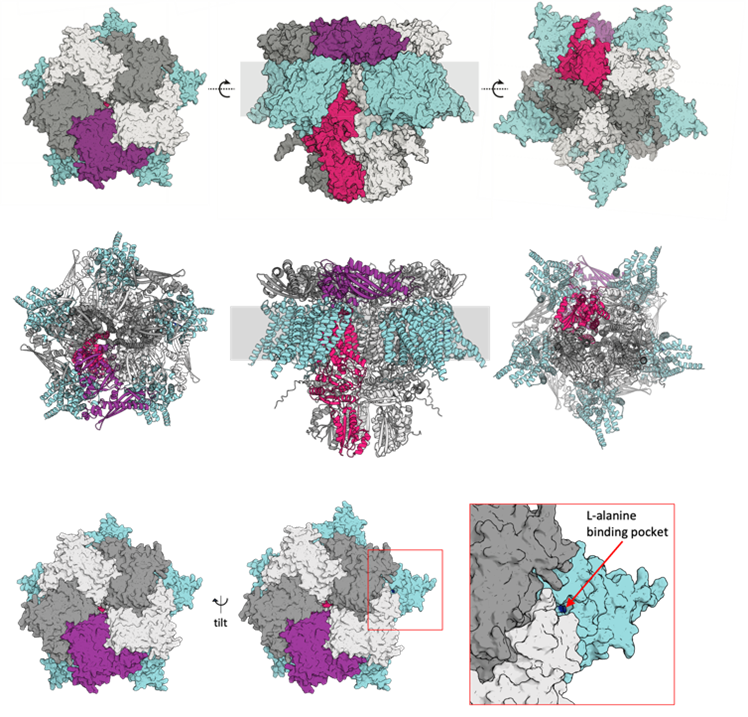
Bacterial spore germination receptors are nutrient-gated ion channels (Science, 2023)
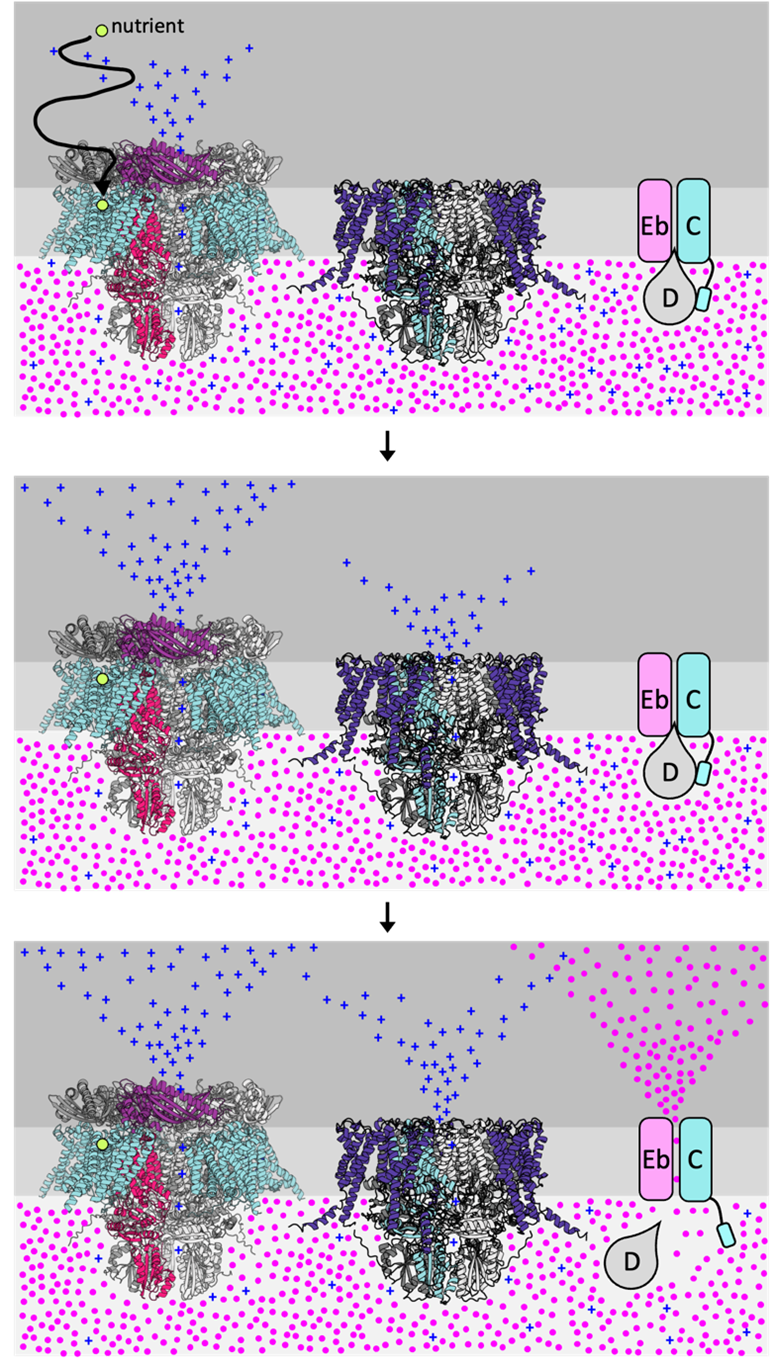
Science, 2023→Genes Dev, 2024→Genes Dev, 2022
The pathway for bacterial spore germination
1. Gao Y, Amon JD, Artzi L, Ramírez-Guadiana FH, Cofsky JC, Kruse AC, Rudner DZ. SpoVAF and FigP form an oligomeric ion channel that amplifies nutrient-triggered spore germination. Genes Dev 38, 31-45 (2024).
2. Gao Y, Amon JD, Artzi L, Ramírez-Guadiana FH, Brock KP, Cofsky JC, Marks DS, Kruse AC, Rudner DZ. Bacterial spore germination receptors are nutrient-gated ion channels. Science 380, 387-391, doi:10.1126/science.adg9829 (2023).
Spotlight by Trends in Microbiology: Spore germination receptors - a new paradigm
3. Gao Y, Barajas-Ornelas RDC, Amon JD, Ramírez-Guadiana FH, Alon A, Brock KP, Marks DS, Kruse AC, Rudner DZ. The SpoVA membrane complex is required for dipicolinic acid import during sporulation and export during germination. Genes Dev 36, 634-646, doi:10.1101/gad.349488.122 (2022).
4. Wenzel M, Celik Gulsoy IN, Gao Y, Teng Z, Willemse J, Middelkamp M, van Rosmalen MGM, Larsen PWB, van der Wel NN, Wuite GJL, Roos WH, Hamoen LW. Control of septum thickness by the curvature of SepF polymers. Proc Natl Acad Sci U S A 118, doi:10.1073/pnas.2002635118 (2021).
5. Syvertsson S, Wang B, Staal J, Gao Y, Kort R, Hamoen LW. Different Resource Allocation in a Bacillus subtilis Population Displaying Bimodal Motility. J Bacteriol 203, e0003721, doi:10.1128/jb.00037-21 (2021).
6. Gao Y, Wenzel M, Jonker MJ, Hamoen LW. Free SepF interferes with recruitment of late cell division proteins. Sci Rep 7, 16928, doi:10.1038/s41598-017-17155-x (2017).
7. Syvertsson S, Vischer NO, Gao Y, Hamoen LW. When Phase Contrast Fails: ChainTracer and NucTracer, Two ImageJ Methods for Semi-Automated Single Cell Analysis Using Membrane or DNA Staining. PLoS One 11, e0151267, doi:10.1371/journal.pone.0151267 (2016).
8. Gao Y, Liu C, Ding Y, Sun C, Zhang R, Xian M, Zhao G. Development of genetically stable Escherichia coli strains for poly(3-hydroxypropionate) production. PLoS One 9, e97845, doi:10.1371/journal.pone.0097845 (2014).
9. Gao Y, Feng X, Xian M, Wang Q, Zhao G. Inducible cell lysis systems in microbial production of bio-based chemicals. Appl Microbiol Biotechnol 97, 7121-7129, doi:10.1007/s00253-013-5100-x (2013).
10. Zeng Y, Guo L, Gao Y, Cui L, Wang M, Huang L, Jiang M, Liu Y, Zhu Y, Xiang H, Li D, Zheng Y. Formation of NifA-PII complex represses ammonium-sensitive nitrogen fixation in diazotrophic proteobacteria lacking NifL. Cell Rep 43(7): 114476 (2024).
11. Zhang X, Liu C, Gao Y, Liu L, Zhang H. The canalization in domesticated yeast: Metabolic traits develop robustness against Hsp90 stress during the evolutionary process. Green Carbon, doi: 10.1016/j.greenca.2024.11.002 (2024).
12. Jiang M, Liu M, Gao Y, Zheng Y. Discovery of a new host for rhizobial symbiont in the ocean. The Innovation Life, 2(3), Article 100077. (2024)
13. Liu C, Li X, Han Y, Li W, Gao Y, Jiang C, Liu Y, Zheng Y. Enhanced pyrite bioleaching through the synergistic interactions between Sulfobacillus thermosulfidooxidans and Alicyclobacillus ferrooxydans. Environ. Technol. Innov. 37, 104005 (2025).