XIANG Hua
Address:NO.1 West Beichen Road, Chaoyang District, Beijing 100101, China
Telephone/fax:010-64807472
E-mail:xiangh@im.ac.cn
Research Interests
Extremophiles: Environmental Microbiome, Genome Editing and Synthetic Biology
(1)Microbiome of Soda Lake and Other Extreme Environments: mechanisms underlying carbon cycling, environmental adaptation, and novel microbial resources for biotechnology;
(2)CRISPR and Genome Editing: CRISPR-Cas and other novel systems in anti-virus, and innovation of genome editing tools;
(3)Reengineering of Halophiles: Mechanism for carbon storage and synthetic biology for bioplastic (PHA) production with halophilic archaea and bacteria.
Education/degrees
B.S. 1991 Biology, Beijing Normal University
M.S. 1994 Genetics, Beijing Normal University
Ph.D 1997 Biochemistry & Molecular biology, Chinese Academy of Medical Sciences & Peking Union Medical College (CAMS&PUMC)
Work experience
Postdoctoral Fellow Training
1997-1999 Dr. H. Tan’s Lab., Institute of Microbiology, Chinese Academy of Sciences, Beijing, China;
1999-2001 Dr. David WC Li’s Lab., University of Medicine & Dentistry of New Jersey (UMDNJ), New Jersy, U.S. A.
Professional Appointments
2001-2003, Principal Investigator and Associate Professor, Institute of Microbiology, Chinese Academy of Sciences (IMCAS)
2003.12-present, Principal Investigator, Professor and Ph.D Advisor, IMCAS
2009, Awardee of the National Distinguished Young Scholar Program, China
2017- present, Director, State Key Laboratory of Microbial Resources
2018—present, Deputy Director-General, IMCAS
2021-present: Secretary-General, Chinese Society for Microbiology
Editorial Board
Journal of Genetics and Genomics (Associate editor/Board member, 2014-)
Frontiers in Microbiology (Associate editor, 2018-)
Frontiers in Genome Editing (Associate Editor, 2019-)
Applied and Environmental Microbiology (Editorial Board member,2013-2018)
mLife (Associate Editor,2021-)
The Innovation (Editorial Board member, 2020-)
Acta Microbiologica Sinica (Board member/ Associate Editor, 2007-)
Acta Laser Biology Sinica (Associate Editor,2018-)
Aquatic Biosystems (Section Editor, 2012-2015)
The main research areas
Molecular Microbiology, Synthetic Biology and Genome Editing
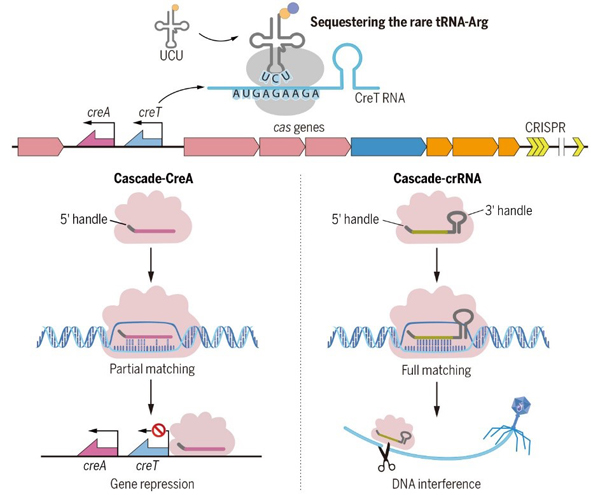
Toxin-antitoxin RNA pairs safeguard CRISPR-Cas systems.
(Science, 2021)
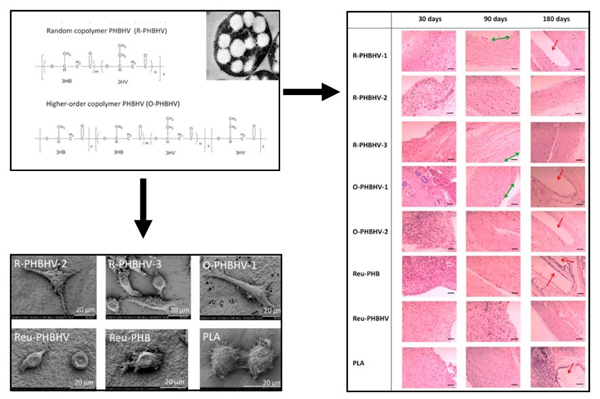
Biosynthesis of tailor-made PHBHV and its superior biocompatibility.
(Biomaterials, 2017)
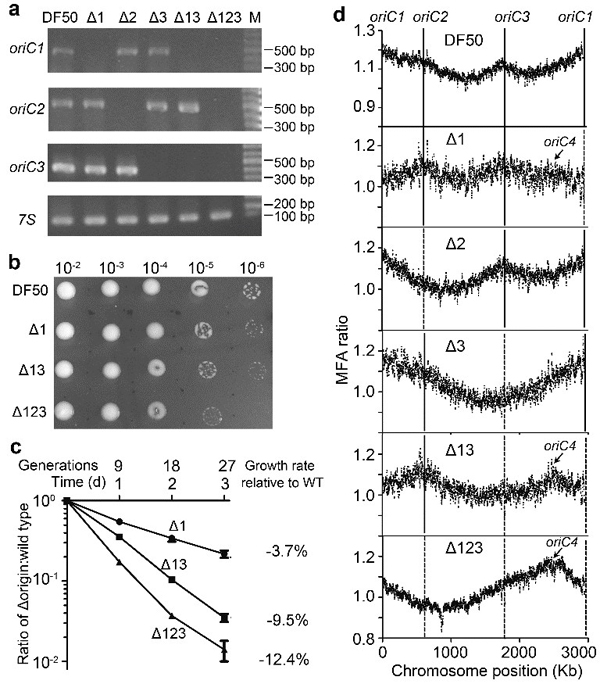
Activation of a dormant replication origin in haloarchaea
(Nature Communications, 2015).

Selected Publication
[1] Zhou H, Zhao D, Zhang S, Xue Q, Zhang M, Yu H, Zhou J, Li M, Kumar S and Xiang H*(2022). Metagenomic insights into the environmental adaptation and metabolism of Candidatus Haloplasmatales, one archaeal order thriving in saline lakes. Environ Microbiol.. doi: 10.1111/1462-2920.15899.
[2] Xue Q, Zhao D, Zhang S, Zhou H, Zuo Z, Zhou J, Li M and Xiang H*(2021). Highly integrated adaptive mechanisms in Spiribacter halalkaliphilus, a bacterium abundant in Chinese soda-saline lakes. Environ Microbiol 23(11): 6463-6482.
[3] Li M, Gong L, Cheng F, Yu H, Zhao D, Wang R, Wang T, Zhang S, Zhou J, Shmakov SA, Koonin EV, Xiang H*.Toxin-antitoxin RNA pairs safeguard CRISPR-Cas systems. Science. 2021; 372(6541): abe5601. Doi: 10.1126/science.abe5601
[4] Lin L, Chen J, Mitra R, Gao Q, Cheng F, Xu T, Zuo Z, Xiang H*, Han J. Optimising PHBV biopolymer production in haloarchaea via CRISPRi-mediated redirection of carbon flux. Commun Biol. 2021;4(1):1007. Doi: 10.1038/s42003-021-02541-z
[5] Cheng F, Wang R, Yu H, Liu C, Yang J, Xiang H*, Li M. Divergent degeneration ofcreAantitoxin genes from minimal CRISPRs and the convergent strategy of tRNA-sequestering CreT toxins. Nucleic Acids Res. 2021;49(18):10677-88. Doi: 10.1093/nar/gkab821
[6] Xu Z, Li M, Li Y, Cao H, Miao L, Xu Z, Higuchi Y, Yamasaki S, Nishino K, Woo PCY, Xiang H*, Yan A. 2019. Native CRISPR-Cas-Mediated Genome Editing Enables Dissecting and Sensitizing Clinical Multidrug-Resistant P. aeruginosa. Cell Rep. 29(6):1707-1717.e3. doi: 10.1016/j.celrep.2019.10.006.
[7] Gong L, Li M, Cheng F, Zhao D, Chen Y, Xiang H*. 2019. Primed adaptation tolerates extensive structural and size variations of the CRISPR RNA guide in Haloarcula hispanica. Nucleic Acids Res.47(11):5880-5891. doi: 10.1093/nar/gkz244.
[8] Li M, Gong L, Zhao D, Zhou J, Xiang H*. 2017. The spacer size of I-B CRISPR is modulated by the terminal sequence of the protospacer. Nucleic Acids Res, 45(8): 4642-4654.
[9] Han J, Wu L, Liu X, Hou J, Zhao L, Chen J, Zhao D, Xiang H*. 2017. Biodegradation and biocompatibility of haloarchaea-produced poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymers. Biomaterials, 139: 172-186.
[10] Wang R, Li M, Gong L, Hu S, Xiang H*. 2016. DNA motifs determining the accuracy of repeat duplication during CRISPR adaptation in Haloarcula hispanica. Nucleic Acids Res, 44(9):4266-77.
[11] Yang H, Wu Z, Liu J, Liu X, Wang L, Cai S, Xiang H*, 2015, Activation of a dormant replication origin is essential for Haloferax mediterranei lacking the primary origins. Nature Communications, 6: 8321.
[12] Liu G, Hou J, Cai S, Zhao D, Cai L, Han J, Zhou J, Xiang H*. 2015. A patatin-like protein associated with the polyhydroxyalkanoate (PHA) granules of Haloferax mediterranei acts as an efficient depolymerase in degradation of native PHA. Appl Environ Microbiol., 81(9):3029-38.
[13] Han J, Wu LP, Hou J, Zhao D, Xiang H*. 2015. Biosynthesis, characterization and hemostasis potential of tailor-made poly(3-hydroxybutyrate-co-3- hydroxyvalerate) produced by Haloferax mediterranei. Biomacromolecules, 16: 578-588
[14] Hou J, Xiang H*, Han J*. 2015. Propionyl coenzyme A (propionyl-CoA) carboxylase in Haloferax mediterranei: Indispensability for propionyl-CoA assimilation and impacts on global metabolism. Appl Environ Microbiol., 81(2):794-804
[15] Cai S, Cai L, Zhao D, Liu G, Han J, Zhou J, Xiang H*. 2015. A novel DNA-binding protein, PhaR, plays a central role in the regulation of polyhydroxyalkanoate accumulation and granule formation in the haloarchaeon Haloferax mediterranei. Appl Environ Microbiol., 81(1):373-385
[16] Li M, Wang R, Xiang H*. 2014. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res, 42(11):7226-35
[17] Cai L, Cai S, Zhao D, Wu J, Wang L, Liu X, Li M, Hou J, Zhou J, Liu J, Han J, Xiang H*. 2014. Analysis of the transcriptional regulator GlpR, promoter elements, and posttranscriptional processing involved in fructose-induced activation of the phosphoenolpyruvate-dependent sugar phosphotransferase system in Haloferax mediterranei. Appl Environ Microbiol., 80(4):1430-40.
[18] Wu Z, Liu J, Yang H, Liu H,Xiang H*.2014. Multiple replication origins with diverse control mechanisms in Haloarcula hispanica. Nucleic Acids Res, 42(4): 2282-2294.
[19] Li M, Wang R, Zhao D,Xiang H*. 2014. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res, 42(4): 2483-2492.
[20] Hou J, Feng B, Han J, Liu H, Zhao D, Zhou J, Xiang H*. 2013. Haloarchaeal type β-ketothiolases involved in poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in Haloferax mediterranei Appl Environ Microbiol., 79(17):5104-11.
[21] Han J, Hou J, Zhang F, Ai G, Li M, Cai S, Liu H, Wang L, Wang Z, Zhang S, Cai L, Zhao D, Zhou J, Xiang H*.2013. Multiple propionyl coenzyme A-supplying pathways for production of the bioplastic poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in Haloferax mediterranei.. Appl Environ Microbiol., 79(9): 2922-31.
[22] Cai S, Cai L, Liu H, Liu X, Han J, Zhou J, Xiang H*. 2012. Identification of the haloarchaeal type phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl Environ Microbiol, 78(6):1946-1952.
[23] Han J, Hou J, Liu H, Cai S, Feng B, Zhou J, Xiang H*. 2010. Wide distribution among halophilic archaea of a novel polyhydroxyalkanoate synthase subtype with homology to bacterial type III synthases. Appl Environ Microbiol, 76(23): 7811-7819
[24] Han J, Lu Q, Zhou L, Liu H, Xiang H*. (2009). Identification of the Polyhydroxyalkanoate (PHA)-Specific Acetoacetyl Coenzyme A Reductase among Multiple FabG Paralogs in Haloarcula hispanica and Reconstruction of the PHA Biosynthetic Pathway in Haloferax volcanii. Appl Environ Microbiol, 75(19): 6168–6175
[25] Lu Q, Han J, Zhou L, Coker JA, DasSarma P, DasSarma S, Xiang H*. 2008. Dissection of the regulatory mechanism of a heat-shock responsive promoter in haloarchaea: a new paradigm for general transcription factor directed archaeal gene regulation. Nucleic Acids Research, 36(9):3031–3042
[26] Pei H, Liu J, Li J, Guo A, Zhou J, Xiang H*. 2007. Mechanism for the TtDnaA-Tt-oriC cooperative interaction at high temperature and duplex opening at an unusual AT-rich region in Thermoanaerobacter tengcongensis. Nucleic Acids Research, 35(9): 3087-3099
[27] Han J, Lu Q, Zhou L, Zhou J, Xiang H*. 2007. Molecular characterization of the phaECHm genes required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui. Applied and Environmental Microbiology, 73(19): 6058-6065
[28] Sun C, Li Y, Mei S, Lu Q, Zhou L, Xiang H*. 2005. A single gene directs both production and immunity of halocin C8 in a haloarchaeal strain AS7092. Molecular Microbiology, 57 (2):537–549