Fungal secondary metabolism plays a pivotal role in pathogen-host interactions, yet the regulatory networks linking metabolic reprogramming to virulence remain largely elusive. Prof. YIN Wenbing’s lab at Institute of Microbiology of Chinese Academy of Sciences found that a novel mechanism exists whereby the RNA-binding protein complex regulates the secondary metabolite Fumiquinazoline C, to thereby mediating the pathogenicity of A. fumigatus. This discovery provides a new post-transcriptional regulation-based theory for the prevention and control of fungal infections. This work was published in Advanced Science.
Invasive aspergillosis, a life-threatening fungal infection primarily caused by the opportunistic pathogen A. fumigatus, represents a significant global health burden and remains a leading cause of mortality. A. fumigatus-produced bioactive secondary metabolites (SMs) play key roles in modulating host immunity and acquiring nutrients, though the regulators and key small molecules that mediate metabolic reprogramming and the association with virulence remain unclear.
Previous studies have demonstrated that fungal SMs are encoded by biosynthetic gene clusters (BGCs) and fine-tuned by a hierarchical regulatory network encompassing pathway-specific regulators, epigenetic modifiers, and global regulators. While RNA-binding proteins (RBPs) are well recognized for their pivotal roles in mediating post-transcriptional modifications in eukaryotes, their functional relevance to fungal metabolic reprogramming and virulence remains largely elusive. In particular, the interplay between RBPs and conserved global regulators, such as LaeB, along with the mechanistic implications of these interactions for BGC activation and fungal pathogenicity, has yet to be elucidated.
This study identifies a conserved regulatory hub in the human pathogen A. fumigatus, where the RNA-binding protein CsdA interacts with the global regulator LaeB in the nucleus to regulate biosynthesis of the secondary metabolite fumiquinazoline C (FqC). Disruption of the CsdA-LaeB interaction hyperactivates FqC production, enhancing fungal colonization and lethality in murine invasive aspergillosis models. Integrative metabolomic and transcriptomic analyses reveal that CsdA and LaeB function as co-regulators of a broader secondary metabolic gene cluster network, with FqC emerging as an effector that mediates virulence in vivo. Genetic validation confirms that FqC is strictly required for the increased virulence phenotype of CsdA- or LaeB-deficient strains, while analyses of clinical isolates demonstrate a striking inverse correlation: reduced CsdA and LaeB expression coincides with elevated FqC production, showing consistency with the infection outcomes of the deletion mutants.
This study indentifies a novel mode of fungal metabolic virulence regulation, providing fresh insights into the post-transcriptional regulatory mechanisms that underlying the link between secondary metabolism and pathogenicity.
The work was supported by the Chinese Academy of Sciences Project for Young Scientists in Basic Research and the Strategic Priority Research Program, the National Natural Science Foundation of China, the National Key Research and Development Program of China.
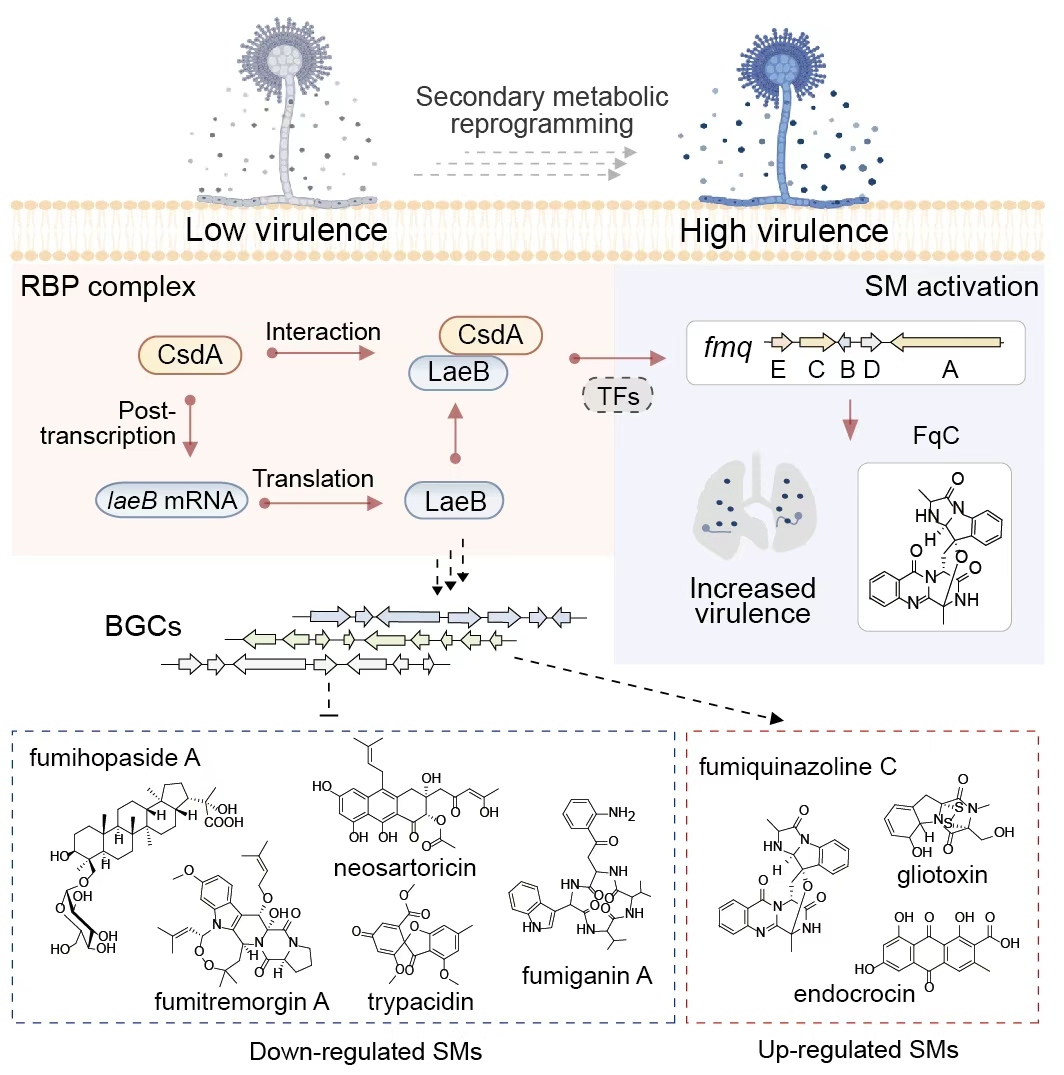
Figure: Model of CsdA-LaeB interaction regulating FqC to drive A. fumigatus virulence. (Image by Prof. YIN Wenbing’s group)
Full text kink: CsdA-LaeB Regulatory Hub Contributes toAspergillus fumigatusVirulence via Fumiquinazoline C Biosynthesis