
Principal Investigator,Professor :
1. In Depth Mining and Design of Gene Editing Tools and Synthetic Biology Catalytic Elements
This research focuses on pioneering innovation in two key components of biomanufacturing: gene editing tools and biocatalytic elements. By integrating microbial multi-omics data and protein structural information, we aim to systematically mine underutilized nuclease systems and high-performance catalytic enzymes from natural sources. Furthermore, we will develop artificial intelligence-based design methods founded on three dimensional structural evolutionary principles and deep learning. These approaches will enhance the functionality of gene editing tools and catalytic elements.
2. Element Interaction Studies and Cell Factory Construction Using Multi Modal AI Models
This research addresses integration challenges in biomanufacturing from individual components to system level performance, with a focus on resolving the critical bottleneck of compatibility between element functions and the cellular chassis. By integrating multi modal data spanning genomics, transcriptomics, and metabolomics, we will construct a virtual microbe AI model capable of simulating microbial physiological states. This model will systematically clarify the synergistic interaction mechanisms and regulatory principles of various elements within metabolic networks.
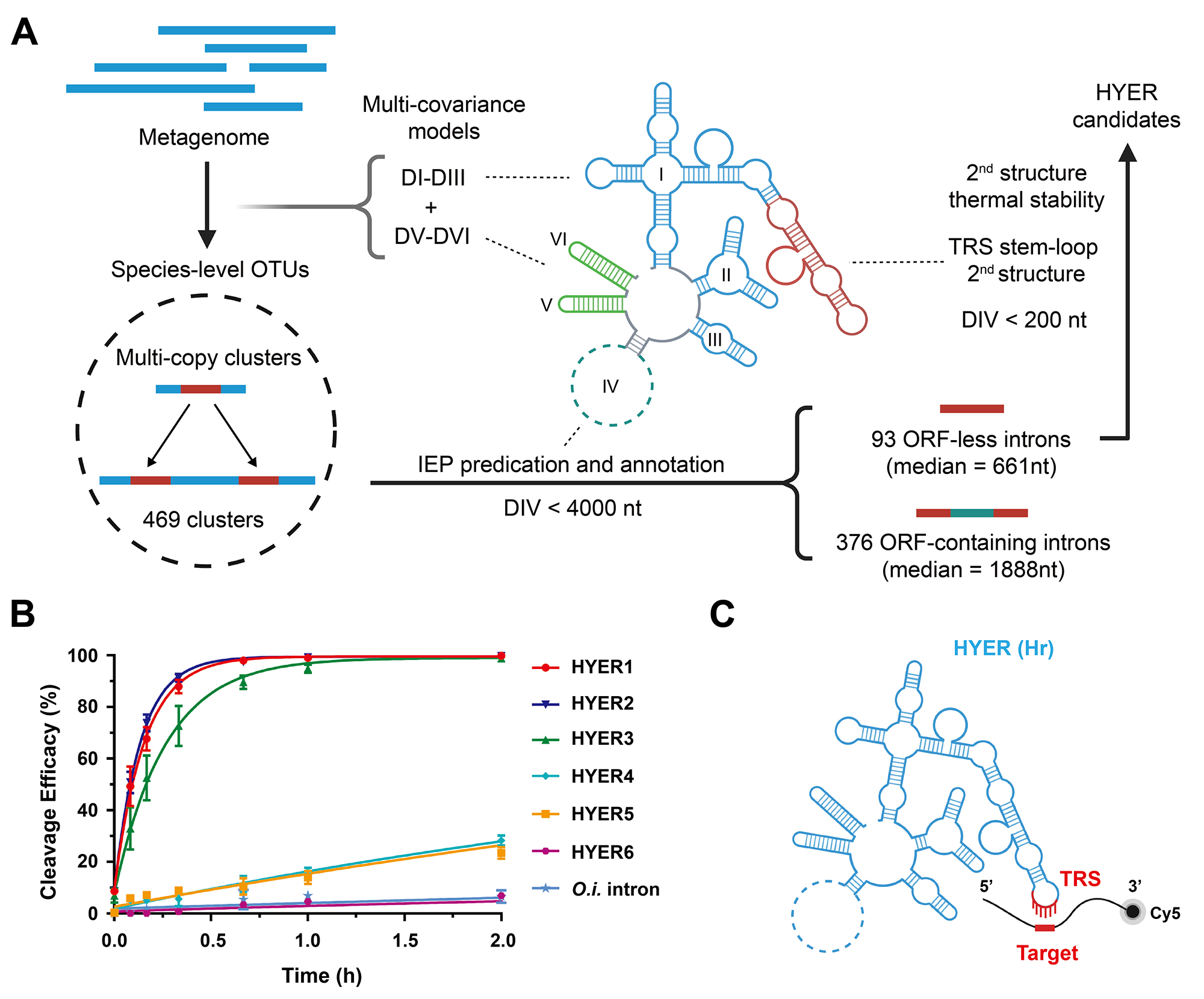
Developed a bioinformatics identification method called "Single-Species Multi-Copy Approach," which successfully identified highly active Group II intron ribozymes in prokaryotic genomes that are composed entirely of RNA elements without any protein components. (Science, 2024, DOI: 10.1126/science.adh4859)
1. S. Zhang, et al. Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity. Nature (2024). (IF = 50.5)
2. Z. Liu#, S. Zhang#, et al. Hydrolytic endonucleolytic ribozyme (HYER) is programmable for sequence-specific DNA cleavage. Science (2024). (IF = 44.7; co-first author)
3. S. Jin#, Z. Zhu#, Y. Li#, S. Zhang#, et al. RNA changes preliminary drive the transformation from transposon to Type V CRISPR system. Cell (2025). (IF = 42.5; co-first author)
4. D. Li#, S. Zhang#, et al. Cas12e homologs evolve variable structural features to facilitate dsDNA cleavage. Nature Communications (2024). (IF = 14.7; co-first author)
5. C. A. Tsuchida#, S. Zhang#, et al. Chimeric CRISPR-CasX enzymes and guide RNAs for improved genome editing activity. Molecular Cell (2022). (IF = 14.5; co-first author)
6. A. Sun#, C. P. Li#, Z. Chen#, S. Zhang#, et al. The compact Casπ (Cas12l) ‘bracelet’ provides a unique structural platform for DNA manipulation. Cell Research (2023). (IF = 28.1; co-first author)